Classical test theory approach to detecting unidirectional and bidirectional (with one crossing location) DIF. This family of statistics is intended for unidimensional tests, and applies a regression-corrected matched-total score approach to quantify the response bias between two or more groups. Can be used for DIF, DBF, and DTF testing with two or more discrete groups.
Usage
SIBTEST(
dat,
group,
suspect_set,
match_set,
focal_name = unique(group)[2],
guess_correction = 0,
Jmin = 5,
na.rm = FALSE,
randomize = FALSE,
C = cbind(1, -diag(length(unique(group)) - 1L)),
pairwise = FALSE,
DIF = FALSE,
p.adjust.method = "none",
permute = 1000,
pk_focal = FALSE,
correction = TRUE,
remove_cross = FALSE,
details = FALSE,
plot = "none",
...
)Arguments
- dat
integer-based dataset to be tested, containing dichotomous or polytomous responses
- group
a (factor) vector indicating group membership with the same length as the number of rows in
dat- suspect_set
an integer vector indicating which items to inspect with SIBTEST. Including only one value will perform a DIF test, while including more than one will perform a simultaneous bundle test (DBF); including all non-matched items will perform DTF. If missing, a simultaneous test using all the items not listed in match_set will be used (i.e., DTF)
- match_set
an integer vector indicating which items to use as the items which are matched (i.e., contain no DIF). These are analogous to 'anchor' items in the likelihood method to locate DIF. If missing, all items other than the items found in the
suspect_setwill be used- focal_name
name of the focal group; e.g.,
'focal'. If not specified then one will be selected automatically usingunique(group)[2]- guess_correction
a vector of numbers from 0 to 1 indicating how much to correct the items for guessing. It's length should be the same as ncol(dat)
- Jmin
the minimum number of observations required when splitting the data into focal and reference groups conditioned on the matched set
- na.rm
logical; remove rows in
datwith any missing values? IfTRUE, rows with missing data will be removed, as well as the corresponding elements in thegroupinput- randomize
logical; perform the crossing test for non-compensatory bias using Li and Stout's (1996) permutation approach? Default is
FALSE, which uses the ad-hoc mixed degrees of freedom method suggested by Chalmers (2018)- C
a contrast matrix to use for pooled testing with more than two groups. Default uses an effects coding approach, where the last group (last column of the matrix) is treated as the reference group, and each column is associated with the respective name via
unique(group)(i.e., the first column is the coefficient forunique(group)[1], second column forunique(group)[2], and so on)- pairwise
logical; perform pairwise comparisons in multi-group applications?
- DIF
logical; should the elements in
suspect_setbe treated one at a time to test for DIF? Use of this logical will treat all other items as part of thematch_setunless this input is provided explicitly. Default isFALSEto allow DBF and DTF tests- p.adjust.method
a character input dictating which
methodto use inp.adjust. when studying more than two groups. Default does not present any p-value adjustments- permute
number of permutations to perform when
randomize = TRUE. Default is 1000- pk_focal
logical; using the group weights from the focal group instead of the total sample? Default is FALSE as per Shealy and Stout's recommendation
- correction
logical; apply the composite correction for the difference between focal composite scores using the true-score regression technique? Default is
TRUE, reflecting Shealy and Stout's linear extrapolation method- remove_cross
logical; remove the subtest information associated with the approximate crossing location? If TRUE this reflects the CSIBTEST definition of Li and Stout (1996); if FALSE, this reflects the version of CSIBTEST utilized by Chalmers (2018). Only applicable in two-group settings (in multi-group this is fixed to FALSE)
- details
logical; return a data.frame containing the details required to compute SIBTEST?
- plot
a character input indicating the type of plot to construct. Options are
'none'(default),'observed'for the scaled focal subtest scores against the matched subtest scores,'weights'for the proportion weights used (i.e., the proportion of observations at each matched score),'difference'for the difference between the scaled focal subtest scores against the matched subtest scores, and'wdifference'for the conditional differences multiplied by each respective weight. Note that the last plot reflects the components used in SIBTEST, and therefore the sum of these plotted observations will equal the beta coefficient for SIBTEST- ...
additional plotting arguments to be passed
Value
a data.frame type object containing the SIBTEST results. Note that
the beta coefficient for (G)CSIBTEST are reported as absolute values to
reflect the sum of the respective area information above and below
the estimated crossing locations
Details
SIBTEST is similar to the Mantel-Haenszel approach for detecting DIF but uses a regression correction based on the KR-20/coefficient alpha reliability index to correct the observed differences when the latent trait distributions are not equal. Function supports the standard SIBTEST for dichotomous and polytomous data (compensatory) and supports crossing DIF testing (i.e., non-compensatory/non-uniform) using the asymptotic sampling distribution version of the Crossing-SIBTEST (CSIBTEST) statistic described by Chalmers (2018) and the permutation method described by Li and Stout (1996). This function also supports the multi-group generalizations (GSIBTEST and GCSIBTEST) proposed by Chalmers and Zheng (2023), where users may specify alternative contrast matrices to evaluate specific comparisons between groups as well as perform joint hypothesis tests.
References
Chalmers, R. P. (2018). Improving the Crossing-SIBTEST statistic for detecting non-uniform DIF. Psychometrika, 83, 2, 376-386.
Chalmers, R., P. (2012). mirt: A Multidimensional Item Response Theory Package for the R Environment. Journal of Statistical Software, 48(6), 1-29. doi:10.18637/jss.v048.i06
Chalmers, R. P. & Zheng, G. (2023). Multi-group Generalizations of SIBTEST and Crossing-SIBTEST. Applied Measurement in Education, 36(2), 171-191, doi:10.1080/08957347.2023.2201703 .
Chang, H. H., Mazzeo, J. & Roussos, L. (1996). DIF for Polytomously Scored Items: An Adaptation of the SIBTEST Procedure. Journal of Educational Measurement, 33, 333-353.
Li, H.-H. & Stout, W. (1996). A new procedure for detection of crossing DIF. Psychometrika, 61, 647-677.
Shealy, R. & Stout, W. (1993). A model-based standardization approach that separates true bias/DIF from group ability differences and detect test bias/DTF as well as item bias/DIF. Psychometrika, 58, 159-194.
Author
Phil Chalmers rphilip.chalmers@gmail.com
Examples
# \donttest{
set.seed(1234)
n <- 30
N <- 500
a <- matrix(1, n)
d <- matrix(rnorm(n), n)
group <- c(rep('reference', N), rep('focal', N*2))
## -------------
# groups completely equal
dat1 <- simdata(a, d, N, itemtype = 'dich')
dat2 <- simdata(a, d, N*2, itemtype = 'dich')
dat <- rbind(dat1, dat2)
# DIF (all other items as anchors)
SIBTEST(dat, group, suspect_set = 6)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.014 0.027 0.265 1 0.606
#> CSIBTEST focal 29 1 0.015 NA 0.331 2 0.848
# Some plots depicting the above tests
SIBTEST(dat, group, suspect_set = 6, plot = 'observed')
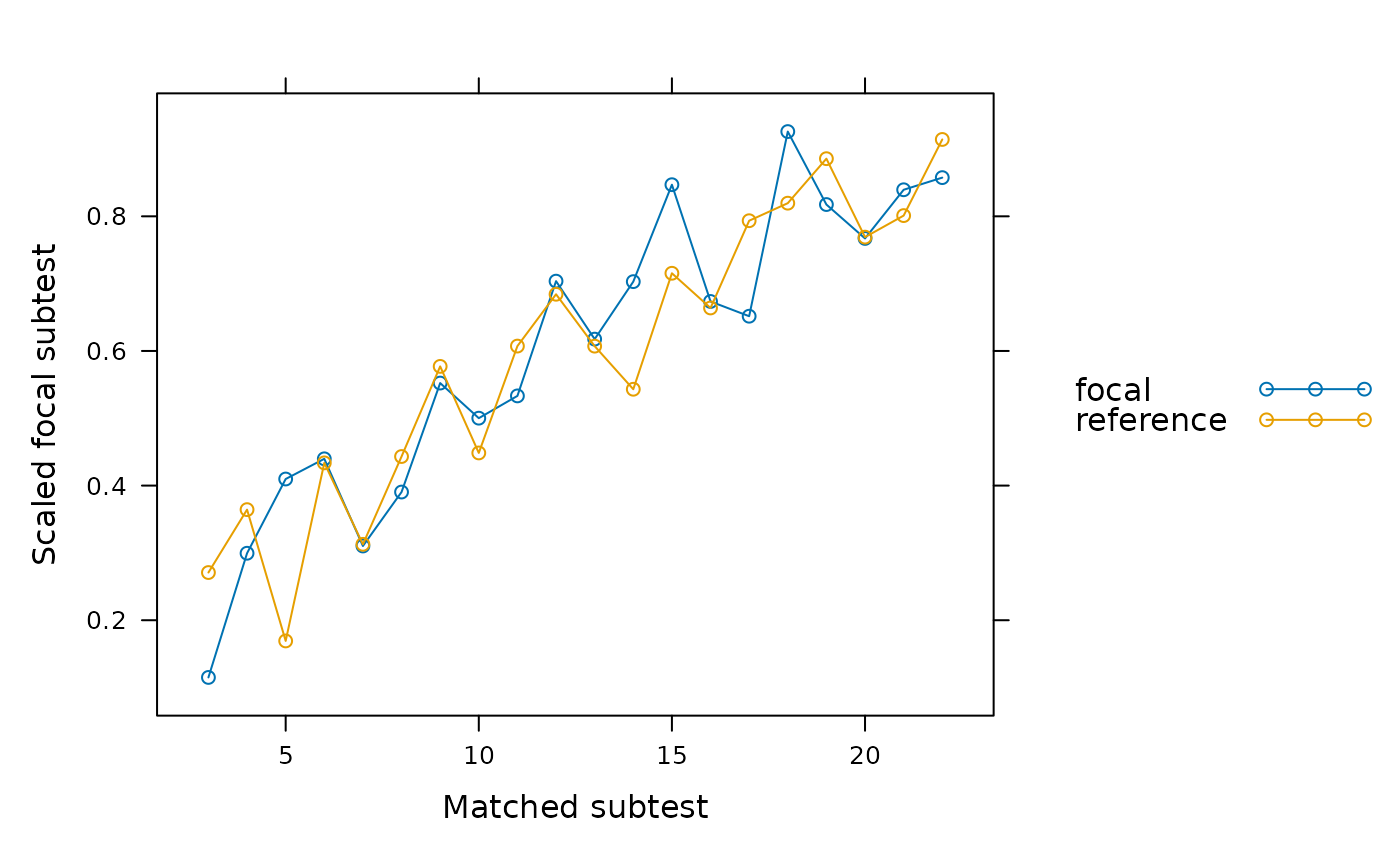 SIBTEST(dat, group, suspect_set = 6, plot = 'weights')
SIBTEST(dat, group, suspect_set = 6, plot = 'weights')
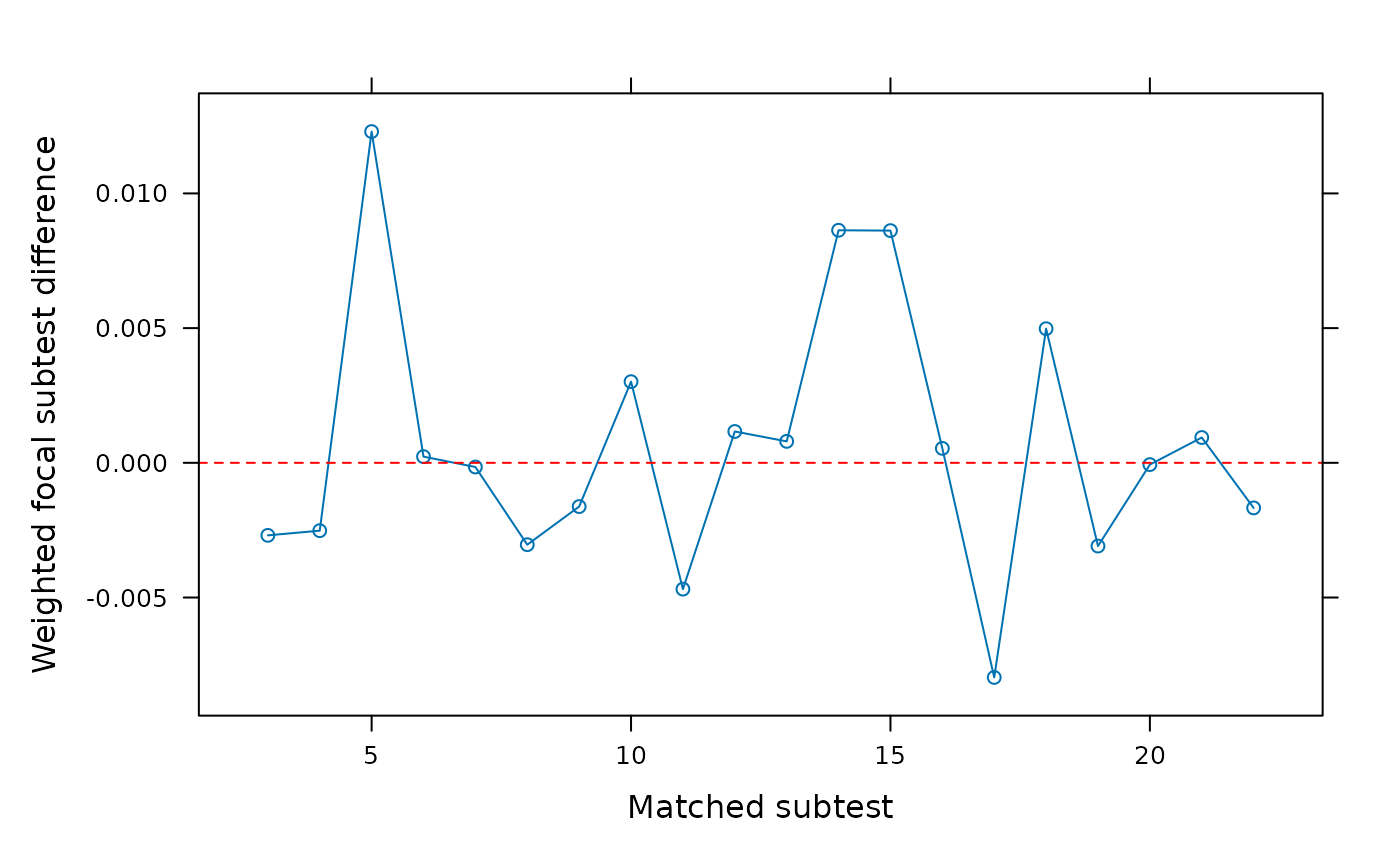
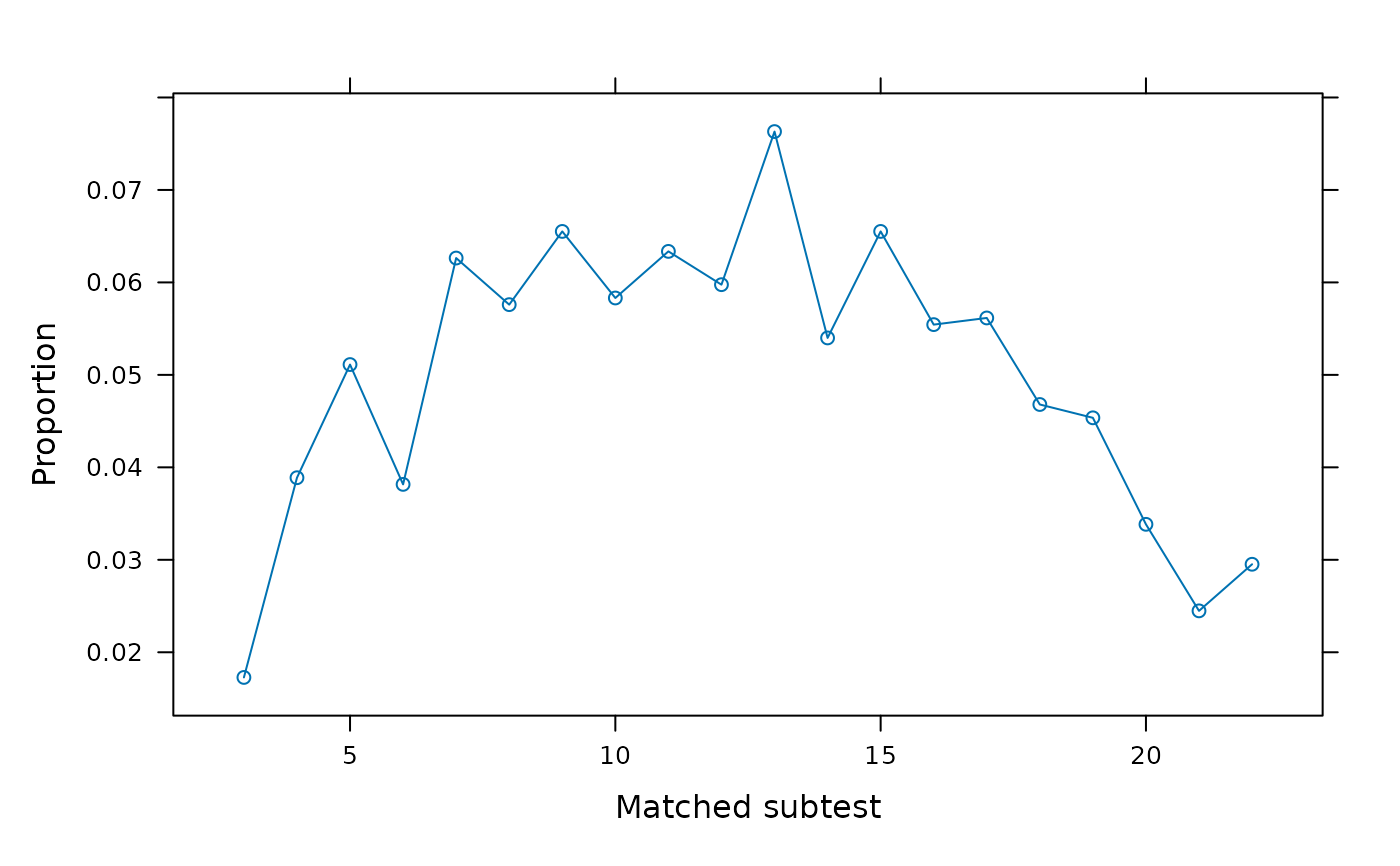 SIBTEST(dat, group, suspect_set = 6, plot = 'wdifference')
SIBTEST(dat, group, suspect_set = 6, plot = 'wdifference')
 # Include CSIBTEST with randomization method
SIBTEST(dat, group, suspect_set = 6, randomize = TRUE)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.014 0.027 0.265 1 0.606
#> CSIBTEST focal 29 1 0.015 0.027 0.827
# remove crossing-location (identical to Li and Stout 1996 definition of CSIBTEST)
SIBTEST(dat, group, suspect_set = 6, randomize = TRUE, remove_cross=TRUE)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.014 0.027 0.265 1 0.606
#> CSIBTEST focal 29 1 0.016 0.026 0.781
# DIF (specific anchors)
SIBTEST(dat, group, match_set = 1:5, suspect_set = 6)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.008 0.028 0.078 1 0.781
#> CSIBTEST focal 5 1 0.004 NA 0.105 2 0.949
SIBTEST(dat, group, match_set = 1:5, suspect_set = 6, randomize=TRUE)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.008 0.028 0.078 1 0.781
#> CSIBTEST focal 5 1 0.004 0.028 1
# DBF (all and specific anchors, respectively)
SIBTEST(dat, group, suspect_set = 11:30)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 10 20 0.053 0.165 0.102 1 0.75
#> CSIBTEST focal 10 20 0.053 NA 0.102 1 0.75
SIBTEST(dat, group, match_set = 1:5, suspect_set = 11:30)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 20 0.166 0.197 0.708 1 0.4
#> CSIBTEST focal 5 20 0.166 NA 0.708 1 0.4
# DTF
SIBTEST(dat, group, suspect_set = 11:30)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 10 20 0.053 0.165 0.102 1 0.75
#> CSIBTEST focal 10 20 0.053 NA 0.102 1 0.75
SIBTEST(dat, group, match_set = 1:10) #equivalent
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 10 20 0.053 0.165 0.102 1 0.75
#> CSIBTEST focal 10 20 0.053 NA 0.102 1 0.75
# different hyper pars
dat1 <- simdata(a, d, N, itemtype = 'dich')
dat2 <- simdata(a, d, N*2, itemtype = 'dich', mu = .5, sigma = matrix(1.5))
dat <- rbind(dat1, dat2)
SIBTEST(dat, group, 6:30)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 25 0.115 0.249 0.214 1 0.644
#> CSIBTEST focal 5 25 0.419 NA 2.966 2 0.227
SIBTEST(dat, group, 11:30)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 10 20 0.098 0.171 0.333 1 0.564
#> CSIBTEST focal 10 20 0.321 NA 3.603 2 0.165
# DIF testing with anchors 1 through 5
SIBTEST(dat, group, 6, match_set = 1:5)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.012 0.027 0.18 1 0.671
#> CSIBTEST focal 5 1 0.030 NA 1.246 2 0.536
SIBTEST(dat, group, 7, match_set = 1:5)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.011 0.029 0.148 1 0.7
#> CSIBTEST focal 5 1 0.038 NA 1.764 2 0.414
SIBTEST(dat, group, 8, match_set = 1:5)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.026 0.028 0.824 1 0.364
#> CSIBTEST focal 5 1 0.026 NA 0.824 1 0.364
# DIF testing with all other items as anchors
SIBTEST(dat, group, 6)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.003 0.027 0.012 1 0.913
#> CSIBTEST focal 29 1 0.014 NA 0.261 2 0.878
SIBTEST(dat, group, 7)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.014 0.028 0.241 1 0.623
#> CSIBTEST focal 29 1 0.032 NA 1.311 2 0.519
SIBTEST(dat, group, 8)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.045 0.026 2.883 1 0.089
#> CSIBTEST focal 29 1 0.045 NA 2.883 1 0.089
## -------------
## systematic differing slopes and intercepts (clear DTF)
dat1 <- simdata(a, d, N, itemtype = 'dich')
dat2 <- simdata(a + c(numeric(15), rnorm(n-15, 1, .25)), d + c(numeric(15), rnorm(n-15, 1, 1)),
N*2, itemtype = 'dich')
dat <- rbind(dat1, dat2)
SIBTEST(dat, group, 6:30)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 25 -2.334 0.264 78.065 1 0
#> CSIBTEST focal 5 25 2.334 NA 78.065 1 0
SIBTEST(dat, group, 11:30)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 10 20 -2.376 0.179 176.396 1 0
#> CSIBTEST focal 10 20 2.376 NA 176.396 1 0
# Some plots depicting the above tests
SIBTEST(dat, group, suspect_set = 11:30, plot = 'observed')
# Include CSIBTEST with randomization method
SIBTEST(dat, group, suspect_set = 6, randomize = TRUE)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.014 0.027 0.265 1 0.606
#> CSIBTEST focal 29 1 0.015 0.027 0.827
# remove crossing-location (identical to Li and Stout 1996 definition of CSIBTEST)
SIBTEST(dat, group, suspect_set = 6, randomize = TRUE, remove_cross=TRUE)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.014 0.027 0.265 1 0.606
#> CSIBTEST focal 29 1 0.016 0.026 0.781
# DIF (specific anchors)
SIBTEST(dat, group, match_set = 1:5, suspect_set = 6)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.008 0.028 0.078 1 0.781
#> CSIBTEST focal 5 1 0.004 NA 0.105 2 0.949
SIBTEST(dat, group, match_set = 1:5, suspect_set = 6, randomize=TRUE)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.008 0.028 0.078 1 0.781
#> CSIBTEST focal 5 1 0.004 0.028 1
# DBF (all and specific anchors, respectively)
SIBTEST(dat, group, suspect_set = 11:30)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 10 20 0.053 0.165 0.102 1 0.75
#> CSIBTEST focal 10 20 0.053 NA 0.102 1 0.75
SIBTEST(dat, group, match_set = 1:5, suspect_set = 11:30)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 20 0.166 0.197 0.708 1 0.4
#> CSIBTEST focal 5 20 0.166 NA 0.708 1 0.4
# DTF
SIBTEST(dat, group, suspect_set = 11:30)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 10 20 0.053 0.165 0.102 1 0.75
#> CSIBTEST focal 10 20 0.053 NA 0.102 1 0.75
SIBTEST(dat, group, match_set = 1:10) #equivalent
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 10 20 0.053 0.165 0.102 1 0.75
#> CSIBTEST focal 10 20 0.053 NA 0.102 1 0.75
# different hyper pars
dat1 <- simdata(a, d, N, itemtype = 'dich')
dat2 <- simdata(a, d, N*2, itemtype = 'dich', mu = .5, sigma = matrix(1.5))
dat <- rbind(dat1, dat2)
SIBTEST(dat, group, 6:30)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 25 0.115 0.249 0.214 1 0.644
#> CSIBTEST focal 5 25 0.419 NA 2.966 2 0.227
SIBTEST(dat, group, 11:30)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 10 20 0.098 0.171 0.333 1 0.564
#> CSIBTEST focal 10 20 0.321 NA 3.603 2 0.165
# DIF testing with anchors 1 through 5
SIBTEST(dat, group, 6, match_set = 1:5)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.012 0.027 0.18 1 0.671
#> CSIBTEST focal 5 1 0.030 NA 1.246 2 0.536
SIBTEST(dat, group, 7, match_set = 1:5)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.011 0.029 0.148 1 0.7
#> CSIBTEST focal 5 1 0.038 NA 1.764 2 0.414
SIBTEST(dat, group, 8, match_set = 1:5)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.026 0.028 0.824 1 0.364
#> CSIBTEST focal 5 1 0.026 NA 0.824 1 0.364
# DIF testing with all other items as anchors
SIBTEST(dat, group, 6)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.003 0.027 0.012 1 0.913
#> CSIBTEST focal 29 1 0.014 NA 0.261 2 0.878
SIBTEST(dat, group, 7)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.014 0.028 0.241 1 0.623
#> CSIBTEST focal 29 1 0.032 NA 1.311 2 0.519
SIBTEST(dat, group, 8)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.045 0.026 2.883 1 0.089
#> CSIBTEST focal 29 1 0.045 NA 2.883 1 0.089
## -------------
## systematic differing slopes and intercepts (clear DTF)
dat1 <- simdata(a, d, N, itemtype = 'dich')
dat2 <- simdata(a + c(numeric(15), rnorm(n-15, 1, .25)), d + c(numeric(15), rnorm(n-15, 1, 1)),
N*2, itemtype = 'dich')
dat <- rbind(dat1, dat2)
SIBTEST(dat, group, 6:30)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 25 -2.334 0.264 78.065 1 0
#> CSIBTEST focal 5 25 2.334 NA 78.065 1 0
SIBTEST(dat, group, 11:30)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 10 20 -2.376 0.179 176.396 1 0
#> CSIBTEST focal 10 20 2.376 NA 176.396 1 0
# Some plots depicting the above tests
SIBTEST(dat, group, suspect_set = 11:30, plot = 'observed')
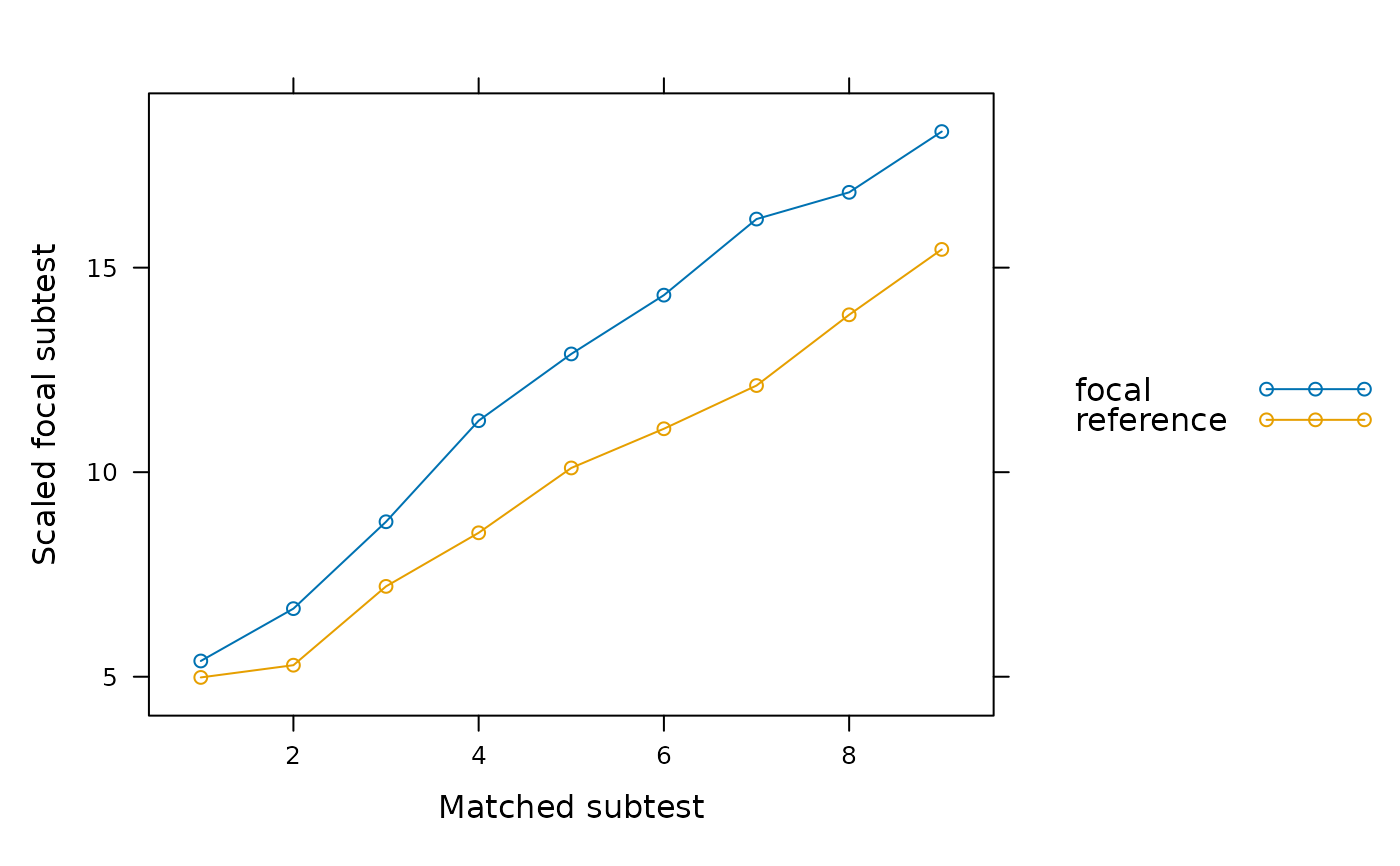 SIBTEST(dat, group, suspect_set = 11:30, plot = 'weights')
SIBTEST(dat, group, suspect_set = 11:30, plot = 'weights')
 SIBTEST(dat, group, suspect_set = 11:30, plot = 'wdifference')
SIBTEST(dat, group, suspect_set = 11:30, plot = 'wdifference')
 # DIF testing using valid anchors
SIBTEST(dat, group, suspect_set = 6, match_set = 1:5)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.013 0.028 0.209 1 0.648
#> CSIBTEST focal 5 1 0.060 NA 5.276 2 0.071
SIBTEST(dat, group, suspect_set = 7, match_set = 1:5)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.011 0.027 0.17 1 0.68
#> CSIBTEST focal 5 1 0.006 NA 0.173 2 0.917
SIBTEST(dat, group, suspect_set = 30, match_set = 1:5)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.335 0.026 166.911 1 0
#> CSIBTEST focal 5 1 0.335 NA 166.911 1 0
# test DIF using specific match_set
SIBTEST(dat, group, suspect_set = 6:30, match_set = 1:5, DIF=TRUE)
#> $Item_6
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.013 0.028 0.209 1 0.648
#> CSIBTEST focal 5 1 0.060 NA 5.276 2 0.071
#>
#> $Item_7
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.011 0.027 0.17 1 0.68
#> CSIBTEST focal 5 1 0.006 NA 0.173 2 0.917
#>
#> $Item_8
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.007 0.028 0.071 1 0.79
#> CSIBTEST focal 5 1 0.007 NA 0.071 1 0.79
#>
#> $Item_9
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.004 0.027 0.022 1 0.882
#> CSIBTEST focal 5 1 0.020 NA 0.562 2 0.755
#>
#> $Item_10
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.031 0.027 1.309 1 0.253
#> CSIBTEST focal 5 1 0.018 NA 1.314 2 0.518
#>
#> $Item_11
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.032 0.028 1.292 1 0.256
#> CSIBTEST focal 5 1 0.032 NA 1.292 1 0.256
#>
#> $Item_12
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.018 0.026 0.448 1 0.503
#> CSIBTEST focal 5 1 0.010 NA 0.457 2 0.796
#>
#> $Item_13
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.000 0.027 0 1 0.99
#> CSIBTEST focal 5 1 0.029 NA 1.135 2 0.567
#>
#> $Item_14
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.004 0.028 0.024 1 0.876
#> CSIBTEST focal 5 1 0.005 NA 0.032 2 0.984
#>
#> $Item_15
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.008 0.027 0.091 1 0.763
#> CSIBTEST focal 5 1 0.012 NA 1.273 2 0.529
#>
#> $Item_16
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.194 0.028 48.949 1 0
#> CSIBTEST focal 5 1 0.194 NA 48.949 1 0
#>
#> $Item_17
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.381 0.026 207.053 1 0
#> CSIBTEST focal 5 1 0.381 NA 207.053 1 0
#>
#> $Item_18
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.122 0.027 20.077 1 0
#> CSIBTEST focal 5 1 0.122 NA 20.077 1 0
#>
#> $Item_19
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.206 0.025 67.462 1 0
#> CSIBTEST focal 5 1 0.206 NA 67.462 1 0
#>
#> $Item_20
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.006 0.018 0.104 1 0.747
#> CSIBTEST focal 5 1 0.010 NA 0.64 2 0.726
#>
#> $Item_21
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.288 0.026 120.661 1 0
#> CSIBTEST focal 5 1 0.288 NA 120.661 1 0
#>
#> $Item_22
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.259 0.027 89.969 1 0
#> CSIBTEST focal 5 1 0.259 NA 89.969 1 0
#>
#> $Item_23
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.235 0.028 72.731 1 0
#> CSIBTEST focal 5 1 0.235 NA 72.731 1 0
#>
#> $Item_24
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.06 0.027 4.759 1 0.029
#> CSIBTEST focal 5 1 0.06 NA 4.759 1 0.029
#>
#> $Item_25
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.38 0.026 209.695 1 0
#> CSIBTEST focal 5 1 0.38 NA 209.695 1 0
#>
#> $Item_26
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.196 0.025 60.776 1 0
#> CSIBTEST focal 5 1 0.196 NA 60.776 1 0
#>
#> $Item_27
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.152 0.027 30.646 1 0
#> CSIBTEST focal 5 1 0.152 NA 30.646 1 0
#>
#> $Item_28
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.311 0.026 138.049 1 0
#> CSIBTEST focal 5 1 0.311 NA 138.049 1 0
#>
#> $Item_29
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.129 0.027 22.567 1 0
#> CSIBTEST focal 5 1 0.129 NA 22.567 1 0
#>
#> $Item_30
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.335 0.026 166.911 1 0
#> CSIBTEST focal 5 1 0.335 NA 166.911 1 0
#>
# test DIF using all-other-as-anchors method (not typically recommended)
SIBTEST(dat, group, suspect_set = 1:30, DIF=TRUE)
#> $Item_1
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.127 0.028 20.939 1 0
#> CSIBTEST focal 29 1 0.127 NA 20.939 1 0
#>
#> $Item_2
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.048 0.029 2.801 1 0.094
#> CSIBTEST focal 29 1 0.089 NA 9.376 2 0.009
#>
#> $Item_3
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.013 0.026 0.25 1 0.617
#> CSIBTEST focal 29 1 0.013 NA 0.25 1 0.617
#>
#> $Item_4
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.067 0.023 8.472 1 0.004
#> CSIBTEST focal 29 1 0.067 NA 8.472 1 0.004
#>
#> $Item_5
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.054 0.028 3.78 1 0.052
#> CSIBTEST focal 29 1 0.071 NA 6.524 2 0.038
#>
#> $Item_6
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.045 0.027 2.839 1 0.092
#> CSIBTEST focal 29 1 0.047 NA 3.679 2 0.159
#>
#> $Item_7
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.082 0.028 8.455 1 0.004
#> CSIBTEST focal 29 1 0.085 NA 9.148 2 0.01
#>
#> $Item_8
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.088 0.028 10.042 1 0.002
#> CSIBTEST focal 29 1 0.108 NA 15.424 2 0
#>
#> $Item_9
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.082 0.029 8.256 1 0.004
#> CSIBTEST focal 29 1 0.099 NA 12.193 2 0.002
#>
#> $Item_10
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.076 0.028 7.264 1 0.007
#> CSIBTEST focal 29 1 0.076 NA 7.264 1 0.007
#>
#> $Item_11
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.096 0.028 12.036 1 0.001
#> CSIBTEST focal 29 1 0.114 NA 17.072 2 0
#>
#> $Item_12
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.105 0.027 14.678 1 0
#> CSIBTEST focal 29 1 0.105 NA 14.678 1 0
#>
#> $Item_13
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.057 0.028 4.098 1 0.043
#> CSIBTEST focal 29 1 0.057 NA 4.098 1 0.043
#>
#> $Item_14
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.082 0.026 9.589 1 0.002
#> CSIBTEST focal 29 1 0.082 NA 9.589 1 0.002
#>
#> $Item_15
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.023 0.026 0.779 1 0.378
#> CSIBTEST focal 29 1 0.023 NA 0.779 1 0.378
#>
#> $Item_16
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.092 0.028 10.83 1 0.001
#> CSIBTEST focal 29 1 0.094 NA 11.373 2 0.003
#>
#> $Item_17
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.328 0.028 137.581 1 0
#> CSIBTEST focal 29 1 0.328 NA 137.581 1 0
#>
#> $Item_18
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.037 0.026 2.017 1 0.155
#> CSIBTEST focal 29 1 0.094 NA 14.977 2 0.001
#>
#> $Item_19
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.315 0.033 93.742 1 0
#> CSIBTEST focal 29 1 0.315 NA 93.742 1 0
#>
#> $Item_20
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.069 0.023 8.926 1 0.003
#> CSIBTEST focal 29 1 0.069 NA 8.926 1 0.003
#>
#> $Item_21
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.225 0.025 80.654 1 0
#> CSIBTEST focal 29 1 0.225 NA 80.654 1 0
#>
#> $Item_22
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.186 0.027 48.41 1 0
#> CSIBTEST focal 29 1 0.186 NA 48.41 1 0
#>
#> $Item_23
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.14 0.028 24.643 1 0
#> CSIBTEST focal 29 1 0.14 NA 24.643 1 0
#>
#> $Item_24
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.146 0.026 30.882 1 0
#> CSIBTEST focal 29 1 0.159 NA 40.395 2 0
#>
#> $Item_25
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.311 0.027 133.904 1 0
#> CSIBTEST focal 29 1 0.311 NA 133.904 1 0
#>
#> $Item_26
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.07 0.025 8.026 1 0.005
#> CSIBTEST focal 29 1 0.07 NA 8.026 1 0.005
#>
#> $Item_27
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.269 0.029 86.816 1 0
#> CSIBTEST focal 29 1 0.269 NA 86.816 1 0
#>
#> $Item_28
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.237 0.027 74.759 1 0
#> CSIBTEST focal 29 1 0.237 NA 74.759 1 0
#>
#> $Item_29
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.036 0.027 1.744 1 0.187
#> CSIBTEST focal 29 1 0.105 NA 14.901 2 0.001
#>
#> $Item_30
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.264 0.028 87.158 1 0
#> CSIBTEST focal 29 1 0.264 NA 87.158 1 0
#>
# randomization method is fairly poor when smaller matched-set used
SIBTEST(dat, group, suspect_set = 30, match_set = 1:5, randomize=TRUE)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.335 0.026 166.911 1 0
#> CSIBTEST focal 5 1 0.335 0.026 0.253
SIBTEST(dat, group, suspect_set = 30, randomize=TRUE)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.264 0.028 87.158 1 0
#> CSIBTEST focal 29 1 0.264 0.028 0
## ----------------------------------
# three group SIBTEST test
set.seed(1234)
n <- 30
N <- 1000
a <- matrix(1, n)
d <- matrix(rnorm(n), n)
group <- c(rep('group1', N), rep('group2', N), rep('group3', N))
# groups completely equal
dat1 <- simdata(a, d, N, itemtype = 'dich')
dat2 <- simdata(a, d, N, itemtype = 'dich')
dat3 <- simdata(a, d, N, itemtype = 'dich')
dat <- rbind(dat1, dat2, dat3)
# omnibus test using effects-coding contrast matrix (default)
SIBTEST(dat, group, suspect_set = 6)
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.005 -0.005 0.084 2 0.959
#> GCSIBTEST 29 1 0.004 0.016 1.027
SIBTEST(dat, group, suspect_set = 6, randomize=TRUE)
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.005 -0.005 0.084 2 0.959
#> GCSIBTEST 29 1 0.004 0.016 1.027 0.847
# explicit contrasts
SIBTEST(dat, group, suspect_set = 6, randomize=TRUE,
C = matrix(c(1,-1,0), 1))
#> n_matched_set n_suspect_set beta X2 df p
#> GSIBTEST 29 1 -0.003 0.02 1 0.888
#> GCSIBTEST 29 1 0.002 0.009 0.957
# test all items for DIF
SIBTEST(dat, group, suspect_set = 1:ncol(dat), DIF=TRUE)
#> $Item_1
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.017 0.001 1.008 2 0.604
#> GCSIBTEST 29 1 0.022 0.031 7.502
#>
#> $Item_2
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.046 -0.022 4.695 2 0.096
#> GCSIBTEST 29 1 0.038 0.026 3.452
#>
#> $Item_3
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.001 -0.002 0.009 2 0.996
#> GCSIBTEST 29 1 0.003 0.008 0.298
#>
#> $Item_4
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.002 -0.005 0.193 2 0.908
#> GCSIBTEST 29 1 0.012 0.006 0.609
#>
#> $Item_5
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.028 0.015 1.807 2 0.405
#> GCSIBTEST 29 1 0.028 0.029 7.164
#>
#> $Item_6
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.005 -0.005 0.084 2 0.959
#> GCSIBTEST 29 1 0.004 0.016 1.027
#>
#> $Item_7
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.007 0.015 1.134 2 0.567
#> GCSIBTEST 29 1 0.012 0.015 0.564
#>
#> $Item_8
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.001 0.007 0.147 2 0.929
#> GCSIBTEST 29 1 0.019 0.011 0.835
#>
#> $Item_9
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.046 0.002 6.347 2 0.042
#> GCSIBTEST 29 1 0.037 0.031 3.719
#>
#> $Item_10
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.004 0.002 0.084 2 0.959
#> GCSIBTEST 29 1 0.036 0.014 3.326
#>
#> $Item_11
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.017 -0.015 0.788 2 0.674
#> GCSIBTEST 29 1 0.017 0.017 2.736
#>
#> $Item_12
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.026 0.024 2.02 2 0.364
#> GCSIBTEST 29 1 0.035 0.023 3.074
#>
#> $Item_13
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.005 0 0.088 2 0.957
#> GCSIBTEST 29 1 0.016 0 0.769
#>
#> $Item_14
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.000 0.005 0.085 2 0.959
#> GCSIBTEST 29 1 0.022 0.031 2.229
#>
#> $Item_15
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.013 0.025 1.558 2 0.459
#> GCSIBTEST 29 1 0.011 0.041 4.631
#>
#> $Item_16
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.003 -0.033 2.934 2 0.231
#> GCSIBTEST 29 1 0.021 0.033 2.469
#>
#> $Item_17
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.001 0.001 0.002 2 0.999
#> GCSIBTEST 29 1 0.002 0.006 0.088
#>
#> $Item_18
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.06 -0.023 8.761 2 0.013
#> GCSIBTEST 29 1 0.06 0.004 10.588
#>
#> $Item_19
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.005 -0.025 1.591 2 0.451
#> GCSIBTEST 29 1 0.029 0.010 4.078
#>
#> $Item_20
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.011 0.002 0.583 2 0.747
#> GCSIBTEST 29 1 0.011 0.014 2.621
#>
#> $Item_21
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.013 0.031 2.247 2 0.325
#> GCSIBTEST 29 1 0.032 0.031 8.87
#>
#> $Item_22
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.010 -0.019 1.868 2 0.393
#> GCSIBTEST 29 1 0.012 0.019 0.792
#>
#> $Item_23
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.007 -0.018 0.745 2 0.689
#> GCSIBTEST 29 1 0.030 0.018 5.375
#>
#> $Item_24
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.006 0.020 0.891 2 0.641
#> GCSIBTEST 29 1 0.016 0.025 3.57
#>
#> $Item_25
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.027 0.015 3.923 2 0.141
#> GCSIBTEST 29 1 0.027 0.019 1.697
#>
#> $Item_26
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.010 0.010 0.434 2 0.805
#> GCSIBTEST 29 1 0.017 0.005 1.637
#>
#> $Item_27
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.023 0.016 1.272 2 0.529
#> GCSIBTEST 29 1 0.023 0.017 1.288
#>
#> $Item_28
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.003 0.003 0.087 2 0.957
#> GCSIBTEST 29 1 0.001 0.016 0.844
#>
#> $Item_29
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.007 -0.027 1.69 2 0.43
#> GCSIBTEST 29 1 0.031 0.027 2.472
#>
#> $Item_30
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.013 -0.011 1.391 2 0.499
#> GCSIBTEST 29 1 0.012 0.020 2.593
#>
SIBTEST(dat, group, suspect_set = 16:ncol(dat), DIF=TRUE,
match_set = 1:15) # specific anchors
#> $Item_16
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 -0.003 -0.035 3.407 2 0.182
#> GCSIBTEST 15 1 0.018 0.035 6.656
#>
#> $Item_17
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 0.000 0.00 0 2 1
#> GCSIBTEST 15 1 0.019 0.03 2.148
#>
#> $Item_18
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 -0.064 -0.027 10.021 2 0.007
#> GCSIBTEST 15 1 0.064 0.010 11.28
#>
#> $Item_19
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 -0.013 -0.029 1.915 2 0.384
#> GCSIBTEST 15 1 0.015 0.027 4.362
#>
#> $Item_20
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 0.01 -0.002 0.817 2 0.665
#> GCSIBTEST 15 1 0.01 0.008 0.54
#>
#> $Item_21
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 0.015 0.022 1.116 2 0.572
#> GCSIBTEST 15 1 0.035 0.022 2.729
#>
#> $Item_22
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 0.005 -0.026 2.51 2 0.285
#> GCSIBTEST 15 1 0.005 0.026 2.51
#>
#> $Item_23
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 -0.008 -0.022 1.108 2 0.575
#> GCSIBTEST 15 1 0.007 0.022 2.045
#>
#> $Item_24
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 0.003 0.016 0.686 2 0.71
#> GCSIBTEST 15 1 0.018 0.023 3.804
#>
#> $Item_25
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 -0.033 0.007 4.161 2 0.125
#> GCSIBTEST 15 1 0.033 0.007 4.161
#>
#> $Item_26
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 0.011 0.007 0.338 2 0.845
#> GCSIBTEST 15 1 0.011 0.005 0.729
#>
#> $Item_27
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 0.021 0.009 0.98 2 0.613
#> GCSIBTEST 15 1 0.023 0.018 1.286
#>
#> $Item_28
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 -0.009 -0.003 0.235 2 0.889
#> GCSIBTEST 15 1 0.035 0.028 10.697
#>
#> $Item_29
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 -0.007 -0.032 2.394 2 0.302
#> GCSIBTEST 15 1 0.018 0.032 5.554
#>
#> $Item_30
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 0.013 -0.012 1.558 2 0.459
#> GCSIBTEST 15 1 0.013 0.028 1.872
#>
# post-hoc between two groups only
pick <- group %in% c('group1', 'group2')
SIBTEST(subset(dat, pick), group[pick], suspect_set = 1:ncol(dat), DIF=TRUE)
#> $Item_1
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.023 0.02 1.399 1 0.237
#> CSIBTEST group2 29 1 0.027 NA 1.95 2 0.377
#>
#> $Item_2
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.045 0.021 4.628 1 0.031
#> CSIBTEST group2 29 1 0.041 NA 4.639 2 0.098
#>
#> $Item_3
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.003 0.021 0.027 1 0.87
#> CSIBTEST group2 29 1 0.003 NA 0.027 1 0.87
#>
#> $Item_4
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.002 0.015 0.018 1 0.894
#> CSIBTEST group2 29 1 0.015 NA 1.744 2 0.418
#>
#> $Item_5
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.03 0.021 1.979 1 0.159
#> CSIBTEST group2 29 1 0.03 NA 1.979 1 0.159
#>
#> $Item_6
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.002 0.021 0.005 1 0.942
#> CSIBTEST group2 29 1 0.003 NA 0.029 2 0.986
#>
#> $Item_7
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.005 0.021 0.06 1 0.807
#> CSIBTEST group2 29 1 0.012 NA 0.342 2 0.843
#>
#> $Item_8
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.002 0.021 0.01 1 0.92
#> CSIBTEST group2 29 1 0.019 NA 1.123 2 0.57
#>
#> $Item_9
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.045 0.021 4.696 1 0.03
#> CSIBTEST group2 29 1 0.035 NA 4.82 2 0.09
#>
#> $Item_10
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.007 0.021 0.121 1 0.728
#> CSIBTEST group2 29 1 0.036 NA 3.296 2 0.192
#>
#> $Item_11
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.017 0.021 0.624 1 0.43
#> CSIBTEST group2 29 1 0.017 NA 0.624 1 0.43
#>
#> $Item_12
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.026 0.02 1.733 1 0.188
#> CSIBTEST group2 29 1 0.037 NA 3.391 2 0.183
#>
#> $Item_13
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.002 0.021 0.009 1 0.925
#> CSIBTEST group2 29 1 0.019 NA 1.139 2 0.566
#>
#> $Item_14
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.001 0.022 0.001 1 0.973
#> CSIBTEST group2 29 1 0.022 NA 1.108 2 0.575
#>
#> $Item_15
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.012 0.02 0.357 1 0.55
#> CSIBTEST group2 29 1 0.010 NA 0.358 2 0.836
#>
#> $Item_16
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.005 0.022 0.064 1 0.801
#> CSIBTEST group2 29 1 0.016 NA 0.547 2 0.761
#>
#> $Item_17
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.002 0.021 0.011 1 0.916
#> CSIBTEST group2 29 1 0.003 NA 0.025 2 0.987
#>
#> $Item_18
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.059 0.02 8.584 1 0.003
#> CSIBTEST group2 29 1 0.059 NA 8.584 1 0.003
#>
#> $Item_19
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.01 0.021 0.248 1 0.619
#> CSIBTEST group2 29 1 0.02 NA 1.031 2 0.597
#>
#> $Item_20
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.018 0.017 1.224 1 0.268
#> CSIBTEST group2 29 1 0.018 NA 1.224 1 0.268
#>
#> $Item_21
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.012 0.021 0.338 1 0.561
#> CSIBTEST group2 29 1 0.030 NA 2.06 2 0.357
#>
#> $Item_22
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.008 0.021 0.141 1 0.707
#> CSIBTEST group2 29 1 0.008 NA 0.171 2 0.918
#>
#> $Item_23
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.007 0.021 0.126 1 0.723
#> CSIBTEST group2 29 1 0.031 NA 2.224 2 0.329
#>
#> $Item_24
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.007 0.022 0.105 1 0.746
#> CSIBTEST group2 29 1 0.012 NA 0.578 2 0.749
#>
#> $Item_25
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.026 0.021 1.468 1 0.226
#> CSIBTEST group2 29 1 0.026 NA 1.468 1 0.226
#>
#> $Item_26
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.009 0.018 0.219 1 0.64
#> CSIBTEST group2 29 1 0.019 NA 3.558 2 0.169
#>
#> $Item_27
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.018 0.021 0.73 1 0.393
#> CSIBTEST group2 29 1 0.018 NA 0.73 1 0.393
#>
#> $Item_28
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.002 0.019 0.015 1 0.903
#> CSIBTEST group2 29 1 0.000 NA 0.017 2 0.991
#>
#> $Item_29
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.010 0.021 0.206 1 0.65
#> CSIBTEST group2 29 1 0.029 NA 1.883 2 0.39
#>
#> $Item_30
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.012 0.02 0.348 1 0.555
#> CSIBTEST group2 29 1 0.012 NA 0.402 2 0.818
#>
# post-hoc pairwise comparison for all groups
SIBTEST(dat, group, suspect_set = 1:ncol(dat), DIF=TRUE, pairwise = TRUE)
#> $group1_group2
#> $group1_group2$Item_1
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.023 0.02 1.399 1 0.237
#> CSIBTEST group2 29 1 0.027 NA 1.95 2 0.377
#>
#> $group1_group2$Item_2
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.045 0.021 4.628 1 0.031
#> CSIBTEST group2 29 1 0.041 NA 4.639 2 0.098
#>
#> $group1_group2$Item_3
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.003 0.021 0.027 1 0.87
#> CSIBTEST group2 29 1 0.003 NA 0.027 1 0.87
#>
#> $group1_group2$Item_4
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.002 0.015 0.018 1 0.894
#> CSIBTEST group2 29 1 0.015 NA 1.744 2 0.418
#>
#> $group1_group2$Item_5
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.03 0.021 1.979 1 0.159
#> CSIBTEST group2 29 1 0.03 NA 1.979 1 0.159
#>
#> $group1_group2$Item_6
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.002 0.021 0.005 1 0.942
#> CSIBTEST group2 29 1 0.003 NA 0.029 2 0.986
#>
#> $group1_group2$Item_7
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.005 0.021 0.06 1 0.807
#> CSIBTEST group2 29 1 0.012 NA 0.342 2 0.843
#>
#> $group1_group2$Item_8
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.002 0.021 0.01 1 0.92
#> CSIBTEST group2 29 1 0.019 NA 1.123 2 0.57
#>
#> $group1_group2$Item_9
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.045 0.021 4.696 1 0.03
#> CSIBTEST group2 29 1 0.035 NA 4.82 2 0.09
#>
#> $group1_group2$Item_10
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.007 0.021 0.121 1 0.728
#> CSIBTEST group2 29 1 0.036 NA 3.296 2 0.192
#>
#> $group1_group2$Item_11
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.017 0.021 0.624 1 0.43
#> CSIBTEST group2 29 1 0.017 NA 0.624 1 0.43
#>
#> $group1_group2$Item_12
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.026 0.02 1.733 1 0.188
#> CSIBTEST group2 29 1 0.037 NA 3.391 2 0.183
#>
#> $group1_group2$Item_13
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.002 0.021 0.009 1 0.925
#> CSIBTEST group2 29 1 0.019 NA 1.139 2 0.566
#>
#> $group1_group2$Item_14
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.001 0.022 0.001 1 0.973
#> CSIBTEST group2 29 1 0.022 NA 1.108 2 0.575
#>
#> $group1_group2$Item_15
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.012 0.02 0.357 1 0.55
#> CSIBTEST group2 29 1 0.010 NA 0.358 2 0.836
#>
#> $group1_group2$Item_16
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.005 0.022 0.064 1 0.801
#> CSIBTEST group2 29 1 0.016 NA 0.547 2 0.761
#>
#> $group1_group2$Item_17
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.002 0.021 0.011 1 0.916
#> CSIBTEST group2 29 1 0.003 NA 0.025 2 0.987
#>
#> $group1_group2$Item_18
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.059 0.02 8.584 1 0.003
#> CSIBTEST group2 29 1 0.059 NA 8.584 1 0.003
#>
#> $group1_group2$Item_19
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.01 0.021 0.248 1 0.619
#> CSIBTEST group2 29 1 0.02 NA 1.031 2 0.597
#>
#> $group1_group2$Item_20
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.018 0.017 1.224 1 0.268
#> CSIBTEST group2 29 1 0.018 NA 1.224 1 0.268
#>
#> $group1_group2$Item_21
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.012 0.021 0.338 1 0.561
#> CSIBTEST group2 29 1 0.030 NA 2.06 2 0.357
#>
#> $group1_group2$Item_22
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.008 0.021 0.141 1 0.707
#> CSIBTEST group2 29 1 0.008 NA 0.171 2 0.918
#>
#> $group1_group2$Item_23
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.007 0.021 0.126 1 0.723
#> CSIBTEST group2 29 1 0.031 NA 2.224 2 0.329
#>
#> $group1_group2$Item_24
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.007 0.022 0.105 1 0.746
#> CSIBTEST group2 29 1 0.012 NA 0.578 2 0.749
#>
#> $group1_group2$Item_25
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.026 0.021 1.468 1 0.226
#> CSIBTEST group2 29 1 0.026 NA 1.468 1 0.226
#>
#> $group1_group2$Item_26
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.009 0.018 0.219 1 0.64
#> CSIBTEST group2 29 1 0.019 NA 3.558 2 0.169
#>
#> $group1_group2$Item_27
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.018 0.021 0.73 1 0.393
#> CSIBTEST group2 29 1 0.018 NA 0.73 1 0.393
#>
#> $group1_group2$Item_28
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.002 0.019 0.015 1 0.903
#> CSIBTEST group2 29 1 0.000 NA 0.017 2 0.991
#>
#> $group1_group2$Item_29
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.010 0.021 0.206 1 0.65
#> CSIBTEST group2 29 1 0.029 NA 1.883 2 0.39
#>
#> $group1_group2$Item_30
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.012 0.02 0.348 1 0.555
#> CSIBTEST group2 29 1 0.012 NA 0.402 2 0.818
#>
#>
#> $group1_group3
#> $group1_group3$Item_1
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.002 0.02 0.006 1 0.937
#> CSIBTEST group3 29 1 0.032 NA 4.561 2 0.102
#>
#> $group1_group3$Item_2
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.022 0.021 1.079 1 0.299
#> CSIBTEST group3 29 1 0.024 NA 1.265 2 0.531
#>
#> $group1_group3$Item_3
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.003 0.02 0.021 1 0.886
#> CSIBTEST group3 29 1 0.009 NA 0.21 2 0.9
#>
#> $group1_group3$Item_4
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.003 0.016 0.043 1 0.836
#> CSIBTEST group3 29 1 0.004 NA 0.079 2 0.961
#>
#> $group1_group3$Item_5
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.019 0.021 0.757 1 0.384
#> CSIBTEST group3 29 1 0.033 NA 2.45 2 0.294
#>
#> $group1_group3$Item_6
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.006 0.021 0.079 1 0.779
#> CSIBTEST group3 29 1 0.017 NA 0.819 2 0.664
#>
#> $group1_group3$Item_7
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.016 0.021 0.589 1 0.443
#> CSIBTEST group3 29 1 0.016 NA 0.589 1 0.443
#>
#> $group1_group3$Item_8
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.007 0.021 0.103 1 0.748
#> CSIBTEST group3 29 1 0.010 NA 0.26 2 0.878
#>
#> $group1_group3$Item_9
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.002 0.021 0.013 1 0.911
#> CSIBTEST group3 29 1 0.032 NA 2.8 2 0.247
#>
#> $group1_group3$Item_10
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.002 0.02 0.01 1 0.919
#> CSIBTEST group3 29 1 0.015 NA 0.974 2 0.615
#>
#> $group1_group3$Item_11
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.014 0.021 0.447 1 0.504
#> CSIBTEST group3 29 1 0.014 NA 1.612 2 0.447
#>
#> $group1_group3$Item_12
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.024 0.02 1.397 1 0.237
#> CSIBTEST group3 29 1 0.023 NA 1.453 2 0.484
#>
#> $group1_group3$Item_13
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.006 0.021 0.092 1 0.762
#> CSIBTEST group3 29 1 0.000 NA 0.126 2 0.939
#>
#> $group1_group3$Item_14
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.004 0.021 0.032 1 0.859
#> CSIBTEST group3 29 1 0.032 NA 2.534 2 0.282
#>
#> $group1_group3$Item_15
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.023 0.02 1.367 1 0.242
#> CSIBTEST group3 29 1 0.040 NA 5.532 2 0.063
#>
#> $group1_group3$Item_16
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.034 0.021 2.489 1 0.115
#> CSIBTEST group3 29 1 0.034 NA 2.489 1 0.115
#>
#> $group1_group3$Item_17
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.000 0.021 0 1 0.985
#> CSIBTEST group3 29 1 0.007 NA 0.141 2 0.932
#>
#> $group1_group3$Item_18
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.022 0.02 1.216 1 0.27
#> CSIBTEST group3 29 1 0.001 NA 2.58 2 0.275
#>
#> $group1_group3$Item_19
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.025 0.02 1.525 1 0.217
#> CSIBTEST group3 29 1 0.010 NA 1.879 2 0.391
#>
#> $group1_group3$Item_20
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.003 0.015 0.034 1 0.854
#> CSIBTEST group3 29 1 0.014 NA 1.113 2 0.573
#>
#> $group1_group3$Item_21
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.031 0.021 2.216 1 0.137
#> CSIBTEST group3 29 1 0.031 NA 2.216 1 0.137
#>
#> $group1_group3$Item_22
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.018 0.021 0.676 1 0.411
#> CSIBTEST group3 29 1 0.018 NA 0.676 1 0.411
#>
#> $group1_group3$Item_23
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.019 0.021 0.823 1 0.364
#> CSIBTEST group3 29 1 0.019 NA 0.823 1 0.364
#>
#> $group1_group3$Item_24
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.018 0.021 0.75 1 0.386
#> CSIBTEST group3 29 1 0.026 NA 1.727 2 0.422
#>
#> $group1_group3$Item_25
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.013 0.021 0.392 1 0.532
#> CSIBTEST group3 29 1 0.018 NA 0.746 2 0.689
#>
#> $group1_group3$Item_26
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.010 0.018 0.304 1 0.581
#> CSIBTEST group3 29 1 0.005 NA 0.325 2 0.85
#>
#> $group1_group3$Item_27
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.017 0.021 0.649 1 0.421
#> CSIBTEST group3 29 1 0.025 NA 1.722 2 0.423
#>
#> $group1_group3$Item_28
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.001 0.02 0.003 1 0.954
#> CSIBTEST group3 29 1 0.009 NA 0.349 2 0.84
#>
#> $group1_group3$Item_29
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.028 0.021 1.764 1 0.184
#> CSIBTEST group3 29 1 0.028 NA 1.764 1 0.184
#>
#> $group1_group3$Item_30
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.011 0.02 0.316 1 0.574
#> CSIBTEST group3 29 1 0.025 NA 1.705 2 0.426
#>
#>
#> $group2_group3
#> $group2_group3$Item_1
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.020 0.019 1.056 1 0.304
#> CSIBTEST group3 29 1 0.003 NA 1.628 2 0.443
#>
#> $group2_group3$Item_2
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.023 0.021 1.233 1 0.267
#> CSIBTEST group3 29 1 0.023 NA 1.233 1 0.267
#>
#> $group2_group3$Item_3
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.000 0.02 0 1 0.993
#> CSIBTEST group3 29 1 0.003 NA 0.122 2 0.941
#>
#> $group2_group3$Item_4
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.005 0.015 0.12 1 0.729
#> CSIBTEST group3 29 1 0.001 NA 0.339 2 0.844
#>
#> $group2_group3$Item_5
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.01 0.021 0.23 1 0.631
#> CSIBTEST group3 29 1 0.00 NA 0.241 2 0.886
#>
#> $group2_group3$Item_6
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.006 0.021 0.078 1 0.779
#> CSIBTEST group3 29 1 0.006 NA 0.078 1 0.779
#>
#> $group2_group3$Item_7
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.019 0.021 0.791 1 0.374
#> CSIBTEST group3 29 1 0.019 NA 0.791 1 0.374
#>
#> $group2_group3$Item_8
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.007 0.021 0.102 1 0.75
#> CSIBTEST group3 29 1 0.013 NA 0.84 2 0.657
#>
#> $group2_group3$Item_9
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.045 0.021 4.764 1 0.029
#> CSIBTEST group3 29 1 0.045 NA 4.764 1 0.029
#>
#> $group2_group3$Item_10
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.008 0.021 0.139 1 0.709
#> CSIBTEST group3 29 1 0.014 NA 0.468 2 0.791
#>
#> $group2_group3$Item_11
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.001 0.021 0.001 1 0.978
#> CSIBTEST group3 29 1 0.017 NA 0.866 2 0.648
#>
#> $group2_group3$Item_12
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.000 0.02 0 1 0.991
#> CSIBTEST group3 29 1 0.002 NA 0.007 2 0.996
#>
#> $group2_group3$Item_13
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.002 0.021 0.014 1 0.907
#> CSIBTEST group3 29 1 0.008 NA 0.204 2 0.903
#>
#> $group2_group3$Item_14
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.005 0.021 0.051 1 0.822
#> CSIBTEST group3 29 1 0.011 NA 1.915 2 0.384
#>
#> $group2_group3$Item_15
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.011 0.02 0.335 1 0.563
#> CSIBTEST group3 29 1 0.049 NA 6.682 2 0.035
#>
#> $group2_group3$Item_16
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.031 0.021 2.131 1 0.144
#> CSIBTEST group3 29 1 0.029 NA 2.194 2 0.334
#>
#> $group2_group3$Item_17
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.004 0.021 0.029 1 0.864
#> CSIBTEST group3 29 1 0.001 NA 0.03 2 0.985
#>
#> $group2_group3$Item_18
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.038 0.021 3.351 1 0.067
#> CSIBTEST group3 29 1 0.038 NA 3.351 1 0.067
#>
#> $group2_group3$Item_19
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.021 0.021 1.06 1 0.303
#> CSIBTEST group3 29 1 0.006 NA 3.523 2 0.172
#>
#> $group2_group3$Item_20
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.017 0.017 0.992 1 0.319
#> CSIBTEST group3 29 1 0.017 NA 0.992 1 0.319
#>
#> $group2_group3$Item_21
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.016 0.021 0.578 1 0.447
#> CSIBTEST group3 29 1 0.034 NA 4.188 2 0.123
#>
#> $group2_group3$Item_22
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.033 0.021 2.372 1 0.124
#> CSIBTEST group3 29 1 0.033 NA 2.372 1 0.124
#>
#> $group2_group3$Item_23
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.009 0.021 0.178 1 0.673
#> CSIBTEST group3 29 1 0.024 NA 1.949 2 0.377
#>
#> $group2_group3$Item_24
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.008 0.022 0.151 1 0.698
#> CSIBTEST group3 29 1 0.018 NA 0.683 2 0.711
#>
#> $group2_group3$Item_25
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.043 0.02 4.495 1 0.034
#> CSIBTEST group3 29 1 0.043 NA 4.495 1 0.034
#>
#> $group2_group3$Item_26
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.003 0.019 0.025 1 0.875
#> CSIBTEST group3 29 1 0.023 NA 3.11 2 0.211
#>
#> $group2_group3$Item_27
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.002 0.021 0.007 1 0.933
#> CSIBTEST group3 29 1 0.013 NA 0.373 2 0.83
#>
#> $group2_group3$Item_28
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.002 0.02 0.013 1 0.909
#> CSIBTEST group3 29 1 0.008 NA 0.354 2 0.838
#>
#> $group2_group3$Item_29
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.018 0.022 0.722 1 0.395
#> CSIBTEST group3 29 1 0.026 NA 1.466 2 0.481
#>
#> $group2_group3$Item_30
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.029 0.02 1.973 1 0.16
#> CSIBTEST group3 29 1 0.032 NA 2.615 2 0.271
#>
#>
## systematic differing slopes and intercepts
dat2 <- simdata(a + c(numeric(15), .5,.5,.5,.5,.5, numeric(10)),
d + c(numeric(15), 0,.6,.7,.8,.9, numeric(10)),
N, itemtype = 'dich')
dat <- rbind(dat1, dat2, dat3)
SIBTEST(dat, group, suspect_set = 16)
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.008 -0.034 4.642 2 0.098
#> GCSIBTEST 29 1 0.059 0.034 8.205
SIBTEST(dat, group, suspect_set = 16, randomize=TRUE)
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.008 -0.034 4.642 2 0.098
#> GCSIBTEST 29 1 0.059 0.034 8.205 0.125
SIBTEST(dat, group, suspect_set = 19)
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.136 -0.025 52.967 2 0
#> GCSIBTEST 29 1 0.130 0.009 61.401
SIBTEST(dat, group, suspect_set = 19, randomize=TRUE)
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.136 -0.025 52.967 2 0
#> GCSIBTEST 29 1 0.130 0.009 61.401 0
SIBTEST(dat, group, suspect_set = c(16, 19), DIF=TRUE)
#> $Item_16
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.008 -0.034 4.642 2 0.098
#> GCSIBTEST 29 1 0.059 0.034 8.205
#>
#> $Item_19
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.136 -0.025 52.967 2 0
#> GCSIBTEST 29 1 0.130 0.009 61.401
#>
SIBTEST(dat, group, suspect_set = c(16, 19), DIF=TRUE, pairwise=TRUE)
#> $group1_group2
#> $group1_group2$Item_16
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.008 0.021 0.154 1 0.695
#> CSIBTEST group2 29 1 0.057 NA 9.536 2 0.008
#>
#> $group1_group2$Item_19
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.138 0.021 45.595 1 0
#> CSIBTEST group2 29 1 0.141 NA 47.776 2 0
#>
#>
#> $group1_group3
#> $group1_group3$Item_16
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.034 0.021 2.489 1 0.115
#> CSIBTEST group3 29 1 0.034 NA 2.489 1 0.115
#>
#> $group1_group3$Item_19
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.025 0.02 1.525 1 0.217
#> CSIBTEST group3 29 1 0.010 NA 1.879 2 0.391
#>
#>
#> $group2_group3
#> $group2_group3$Item_16
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.039 0.02 3.721 1 0.054
#> CSIBTEST group3 29 1 0.040 NA 4.185 2 0.123
#>
#> $group2_group3$Item_19
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.112 0.02 30.503 1 0
#> CSIBTEST group3 29 1 0.118 NA 37.348 2 0
#>
#>
# }
# DIF testing using valid anchors
SIBTEST(dat, group, suspect_set = 6, match_set = 1:5)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.013 0.028 0.209 1 0.648
#> CSIBTEST focal 5 1 0.060 NA 5.276 2 0.071
SIBTEST(dat, group, suspect_set = 7, match_set = 1:5)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.011 0.027 0.17 1 0.68
#> CSIBTEST focal 5 1 0.006 NA 0.173 2 0.917
SIBTEST(dat, group, suspect_set = 30, match_set = 1:5)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.335 0.026 166.911 1 0
#> CSIBTEST focal 5 1 0.335 NA 166.911 1 0
# test DIF using specific match_set
SIBTEST(dat, group, suspect_set = 6:30, match_set = 1:5, DIF=TRUE)
#> $Item_6
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.013 0.028 0.209 1 0.648
#> CSIBTEST focal 5 1 0.060 NA 5.276 2 0.071
#>
#> $Item_7
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.011 0.027 0.17 1 0.68
#> CSIBTEST focal 5 1 0.006 NA 0.173 2 0.917
#>
#> $Item_8
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.007 0.028 0.071 1 0.79
#> CSIBTEST focal 5 1 0.007 NA 0.071 1 0.79
#>
#> $Item_9
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.004 0.027 0.022 1 0.882
#> CSIBTEST focal 5 1 0.020 NA 0.562 2 0.755
#>
#> $Item_10
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.031 0.027 1.309 1 0.253
#> CSIBTEST focal 5 1 0.018 NA 1.314 2 0.518
#>
#> $Item_11
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.032 0.028 1.292 1 0.256
#> CSIBTEST focal 5 1 0.032 NA 1.292 1 0.256
#>
#> $Item_12
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.018 0.026 0.448 1 0.503
#> CSIBTEST focal 5 1 0.010 NA 0.457 2 0.796
#>
#> $Item_13
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.000 0.027 0 1 0.99
#> CSIBTEST focal 5 1 0.029 NA 1.135 2 0.567
#>
#> $Item_14
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.004 0.028 0.024 1 0.876
#> CSIBTEST focal 5 1 0.005 NA 0.032 2 0.984
#>
#> $Item_15
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.008 0.027 0.091 1 0.763
#> CSIBTEST focal 5 1 0.012 NA 1.273 2 0.529
#>
#> $Item_16
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.194 0.028 48.949 1 0
#> CSIBTEST focal 5 1 0.194 NA 48.949 1 0
#>
#> $Item_17
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.381 0.026 207.053 1 0
#> CSIBTEST focal 5 1 0.381 NA 207.053 1 0
#>
#> $Item_18
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.122 0.027 20.077 1 0
#> CSIBTEST focal 5 1 0.122 NA 20.077 1 0
#>
#> $Item_19
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.206 0.025 67.462 1 0
#> CSIBTEST focal 5 1 0.206 NA 67.462 1 0
#>
#> $Item_20
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.006 0.018 0.104 1 0.747
#> CSIBTEST focal 5 1 0.010 NA 0.64 2 0.726
#>
#> $Item_21
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.288 0.026 120.661 1 0
#> CSIBTEST focal 5 1 0.288 NA 120.661 1 0
#>
#> $Item_22
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.259 0.027 89.969 1 0
#> CSIBTEST focal 5 1 0.259 NA 89.969 1 0
#>
#> $Item_23
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.235 0.028 72.731 1 0
#> CSIBTEST focal 5 1 0.235 NA 72.731 1 0
#>
#> $Item_24
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.06 0.027 4.759 1 0.029
#> CSIBTEST focal 5 1 0.06 NA 4.759 1 0.029
#>
#> $Item_25
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.38 0.026 209.695 1 0
#> CSIBTEST focal 5 1 0.38 NA 209.695 1 0
#>
#> $Item_26
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.196 0.025 60.776 1 0
#> CSIBTEST focal 5 1 0.196 NA 60.776 1 0
#>
#> $Item_27
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 0.152 0.027 30.646 1 0
#> CSIBTEST focal 5 1 0.152 NA 30.646 1 0
#>
#> $Item_28
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.311 0.026 138.049 1 0
#> CSIBTEST focal 5 1 0.311 NA 138.049 1 0
#>
#> $Item_29
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.129 0.027 22.567 1 0
#> CSIBTEST focal 5 1 0.129 NA 22.567 1 0
#>
#> $Item_30
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.335 0.026 166.911 1 0
#> CSIBTEST focal 5 1 0.335 NA 166.911 1 0
#>
# test DIF using all-other-as-anchors method (not typically recommended)
SIBTEST(dat, group, suspect_set = 1:30, DIF=TRUE)
#> $Item_1
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.127 0.028 20.939 1 0
#> CSIBTEST focal 29 1 0.127 NA 20.939 1 0
#>
#> $Item_2
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.048 0.029 2.801 1 0.094
#> CSIBTEST focal 29 1 0.089 NA 9.376 2 0.009
#>
#> $Item_3
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.013 0.026 0.25 1 0.617
#> CSIBTEST focal 29 1 0.013 NA 0.25 1 0.617
#>
#> $Item_4
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.067 0.023 8.472 1 0.004
#> CSIBTEST focal 29 1 0.067 NA 8.472 1 0.004
#>
#> $Item_5
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.054 0.028 3.78 1 0.052
#> CSIBTEST focal 29 1 0.071 NA 6.524 2 0.038
#>
#> $Item_6
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.045 0.027 2.839 1 0.092
#> CSIBTEST focal 29 1 0.047 NA 3.679 2 0.159
#>
#> $Item_7
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.082 0.028 8.455 1 0.004
#> CSIBTEST focal 29 1 0.085 NA 9.148 2 0.01
#>
#> $Item_8
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.088 0.028 10.042 1 0.002
#> CSIBTEST focal 29 1 0.108 NA 15.424 2 0
#>
#> $Item_9
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.082 0.029 8.256 1 0.004
#> CSIBTEST focal 29 1 0.099 NA 12.193 2 0.002
#>
#> $Item_10
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.076 0.028 7.264 1 0.007
#> CSIBTEST focal 29 1 0.076 NA 7.264 1 0.007
#>
#> $Item_11
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.096 0.028 12.036 1 0.001
#> CSIBTEST focal 29 1 0.114 NA 17.072 2 0
#>
#> $Item_12
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.105 0.027 14.678 1 0
#> CSIBTEST focal 29 1 0.105 NA 14.678 1 0
#>
#> $Item_13
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.057 0.028 4.098 1 0.043
#> CSIBTEST focal 29 1 0.057 NA 4.098 1 0.043
#>
#> $Item_14
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.082 0.026 9.589 1 0.002
#> CSIBTEST focal 29 1 0.082 NA 9.589 1 0.002
#>
#> $Item_15
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.023 0.026 0.779 1 0.378
#> CSIBTEST focal 29 1 0.023 NA 0.779 1 0.378
#>
#> $Item_16
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.092 0.028 10.83 1 0.001
#> CSIBTEST focal 29 1 0.094 NA 11.373 2 0.003
#>
#> $Item_17
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.328 0.028 137.581 1 0
#> CSIBTEST focal 29 1 0.328 NA 137.581 1 0
#>
#> $Item_18
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.037 0.026 2.017 1 0.155
#> CSIBTEST focal 29 1 0.094 NA 14.977 2 0.001
#>
#> $Item_19
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.315 0.033 93.742 1 0
#> CSIBTEST focal 29 1 0.315 NA 93.742 1 0
#>
#> $Item_20
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.069 0.023 8.926 1 0.003
#> CSIBTEST focal 29 1 0.069 NA 8.926 1 0.003
#>
#> $Item_21
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.225 0.025 80.654 1 0
#> CSIBTEST focal 29 1 0.225 NA 80.654 1 0
#>
#> $Item_22
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.186 0.027 48.41 1 0
#> CSIBTEST focal 29 1 0.186 NA 48.41 1 0
#>
#> $Item_23
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.14 0.028 24.643 1 0
#> CSIBTEST focal 29 1 0.14 NA 24.643 1 0
#>
#> $Item_24
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.146 0.026 30.882 1 0
#> CSIBTEST focal 29 1 0.159 NA 40.395 2 0
#>
#> $Item_25
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.311 0.027 133.904 1 0
#> CSIBTEST focal 29 1 0.311 NA 133.904 1 0
#>
#> $Item_26
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.07 0.025 8.026 1 0.005
#> CSIBTEST focal 29 1 0.07 NA 8.026 1 0.005
#>
#> $Item_27
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 0.269 0.029 86.816 1 0
#> CSIBTEST focal 29 1 0.269 NA 86.816 1 0
#>
#> $Item_28
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.237 0.027 74.759 1 0
#> CSIBTEST focal 29 1 0.237 NA 74.759 1 0
#>
#> $Item_29
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.036 0.027 1.744 1 0.187
#> CSIBTEST focal 29 1 0.105 NA 14.901 2 0.001
#>
#> $Item_30
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.264 0.028 87.158 1 0
#> CSIBTEST focal 29 1 0.264 NA 87.158 1 0
#>
# randomization method is fairly poor when smaller matched-set used
SIBTEST(dat, group, suspect_set = 30, match_set = 1:5, randomize=TRUE)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 5 1 -0.335 0.026 166.911 1 0
#> CSIBTEST focal 5 1 0.335 0.026 0.253
SIBTEST(dat, group, suspect_set = 30, randomize=TRUE)
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST focal 29 1 -0.264 0.028 87.158 1 0
#> CSIBTEST focal 29 1 0.264 0.028 0
## ----------------------------------
# three group SIBTEST test
set.seed(1234)
n <- 30
N <- 1000
a <- matrix(1, n)
d <- matrix(rnorm(n), n)
group <- c(rep('group1', N), rep('group2', N), rep('group3', N))
# groups completely equal
dat1 <- simdata(a, d, N, itemtype = 'dich')
dat2 <- simdata(a, d, N, itemtype = 'dich')
dat3 <- simdata(a, d, N, itemtype = 'dich')
dat <- rbind(dat1, dat2, dat3)
# omnibus test using effects-coding contrast matrix (default)
SIBTEST(dat, group, suspect_set = 6)
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.005 -0.005 0.084 2 0.959
#> GCSIBTEST 29 1 0.004 0.016 1.027
SIBTEST(dat, group, suspect_set = 6, randomize=TRUE)
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.005 -0.005 0.084 2 0.959
#> GCSIBTEST 29 1 0.004 0.016 1.027 0.847
# explicit contrasts
SIBTEST(dat, group, suspect_set = 6, randomize=TRUE,
C = matrix(c(1,-1,0), 1))
#> n_matched_set n_suspect_set beta X2 df p
#> GSIBTEST 29 1 -0.003 0.02 1 0.888
#> GCSIBTEST 29 1 0.002 0.009 0.957
# test all items for DIF
SIBTEST(dat, group, suspect_set = 1:ncol(dat), DIF=TRUE)
#> $Item_1
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.017 0.001 1.008 2 0.604
#> GCSIBTEST 29 1 0.022 0.031 7.502
#>
#> $Item_2
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.046 -0.022 4.695 2 0.096
#> GCSIBTEST 29 1 0.038 0.026 3.452
#>
#> $Item_3
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.001 -0.002 0.009 2 0.996
#> GCSIBTEST 29 1 0.003 0.008 0.298
#>
#> $Item_4
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.002 -0.005 0.193 2 0.908
#> GCSIBTEST 29 1 0.012 0.006 0.609
#>
#> $Item_5
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.028 0.015 1.807 2 0.405
#> GCSIBTEST 29 1 0.028 0.029 7.164
#>
#> $Item_6
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.005 -0.005 0.084 2 0.959
#> GCSIBTEST 29 1 0.004 0.016 1.027
#>
#> $Item_7
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.007 0.015 1.134 2 0.567
#> GCSIBTEST 29 1 0.012 0.015 0.564
#>
#> $Item_8
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.001 0.007 0.147 2 0.929
#> GCSIBTEST 29 1 0.019 0.011 0.835
#>
#> $Item_9
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.046 0.002 6.347 2 0.042
#> GCSIBTEST 29 1 0.037 0.031 3.719
#>
#> $Item_10
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.004 0.002 0.084 2 0.959
#> GCSIBTEST 29 1 0.036 0.014 3.326
#>
#> $Item_11
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.017 -0.015 0.788 2 0.674
#> GCSIBTEST 29 1 0.017 0.017 2.736
#>
#> $Item_12
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.026 0.024 2.02 2 0.364
#> GCSIBTEST 29 1 0.035 0.023 3.074
#>
#> $Item_13
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.005 0 0.088 2 0.957
#> GCSIBTEST 29 1 0.016 0 0.769
#>
#> $Item_14
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.000 0.005 0.085 2 0.959
#> GCSIBTEST 29 1 0.022 0.031 2.229
#>
#> $Item_15
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.013 0.025 1.558 2 0.459
#> GCSIBTEST 29 1 0.011 0.041 4.631
#>
#> $Item_16
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.003 -0.033 2.934 2 0.231
#> GCSIBTEST 29 1 0.021 0.033 2.469
#>
#> $Item_17
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.001 0.001 0.002 2 0.999
#> GCSIBTEST 29 1 0.002 0.006 0.088
#>
#> $Item_18
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.06 -0.023 8.761 2 0.013
#> GCSIBTEST 29 1 0.06 0.004 10.588
#>
#> $Item_19
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.005 -0.025 1.591 2 0.451
#> GCSIBTEST 29 1 0.029 0.010 4.078
#>
#> $Item_20
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.011 0.002 0.583 2 0.747
#> GCSIBTEST 29 1 0.011 0.014 2.621
#>
#> $Item_21
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.013 0.031 2.247 2 0.325
#> GCSIBTEST 29 1 0.032 0.031 8.87
#>
#> $Item_22
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.010 -0.019 1.868 2 0.393
#> GCSIBTEST 29 1 0.012 0.019 0.792
#>
#> $Item_23
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.007 -0.018 0.745 2 0.689
#> GCSIBTEST 29 1 0.030 0.018 5.375
#>
#> $Item_24
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.006 0.020 0.891 2 0.641
#> GCSIBTEST 29 1 0.016 0.025 3.57
#>
#> $Item_25
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.027 0.015 3.923 2 0.141
#> GCSIBTEST 29 1 0.027 0.019 1.697
#>
#> $Item_26
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.010 0.010 0.434 2 0.805
#> GCSIBTEST 29 1 0.017 0.005 1.637
#>
#> $Item_27
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.023 0.016 1.272 2 0.529
#> GCSIBTEST 29 1 0.023 0.017 1.288
#>
#> $Item_28
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.003 0.003 0.087 2 0.957
#> GCSIBTEST 29 1 0.001 0.016 0.844
#>
#> $Item_29
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.007 -0.027 1.69 2 0.43
#> GCSIBTEST 29 1 0.031 0.027 2.472
#>
#> $Item_30
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.013 -0.011 1.391 2 0.499
#> GCSIBTEST 29 1 0.012 0.020 2.593
#>
SIBTEST(dat, group, suspect_set = 16:ncol(dat), DIF=TRUE,
match_set = 1:15) # specific anchors
#> $Item_16
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 -0.003 -0.035 3.407 2 0.182
#> GCSIBTEST 15 1 0.018 0.035 6.656
#>
#> $Item_17
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 0.000 0.00 0 2 1
#> GCSIBTEST 15 1 0.019 0.03 2.148
#>
#> $Item_18
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 -0.064 -0.027 10.021 2 0.007
#> GCSIBTEST 15 1 0.064 0.010 11.28
#>
#> $Item_19
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 -0.013 -0.029 1.915 2 0.384
#> GCSIBTEST 15 1 0.015 0.027 4.362
#>
#> $Item_20
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 0.01 -0.002 0.817 2 0.665
#> GCSIBTEST 15 1 0.01 0.008 0.54
#>
#> $Item_21
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 0.015 0.022 1.116 2 0.572
#> GCSIBTEST 15 1 0.035 0.022 2.729
#>
#> $Item_22
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 0.005 -0.026 2.51 2 0.285
#> GCSIBTEST 15 1 0.005 0.026 2.51
#>
#> $Item_23
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 -0.008 -0.022 1.108 2 0.575
#> GCSIBTEST 15 1 0.007 0.022 2.045
#>
#> $Item_24
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 0.003 0.016 0.686 2 0.71
#> GCSIBTEST 15 1 0.018 0.023 3.804
#>
#> $Item_25
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 -0.033 0.007 4.161 2 0.125
#> GCSIBTEST 15 1 0.033 0.007 4.161
#>
#> $Item_26
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 0.011 0.007 0.338 2 0.845
#> GCSIBTEST 15 1 0.011 0.005 0.729
#>
#> $Item_27
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 0.021 0.009 0.98 2 0.613
#> GCSIBTEST 15 1 0.023 0.018 1.286
#>
#> $Item_28
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 -0.009 -0.003 0.235 2 0.889
#> GCSIBTEST 15 1 0.035 0.028 10.697
#>
#> $Item_29
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 -0.007 -0.032 2.394 2 0.302
#> GCSIBTEST 15 1 0.018 0.032 5.554
#>
#> $Item_30
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 15 1 0.013 -0.012 1.558 2 0.459
#> GCSIBTEST 15 1 0.013 0.028 1.872
#>
# post-hoc between two groups only
pick <- group %in% c('group1', 'group2')
SIBTEST(subset(dat, pick), group[pick], suspect_set = 1:ncol(dat), DIF=TRUE)
#> $Item_1
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.023 0.02 1.399 1 0.237
#> CSIBTEST group2 29 1 0.027 NA 1.95 2 0.377
#>
#> $Item_2
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.045 0.021 4.628 1 0.031
#> CSIBTEST group2 29 1 0.041 NA 4.639 2 0.098
#>
#> $Item_3
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.003 0.021 0.027 1 0.87
#> CSIBTEST group2 29 1 0.003 NA 0.027 1 0.87
#>
#> $Item_4
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.002 0.015 0.018 1 0.894
#> CSIBTEST group2 29 1 0.015 NA 1.744 2 0.418
#>
#> $Item_5
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.03 0.021 1.979 1 0.159
#> CSIBTEST group2 29 1 0.03 NA 1.979 1 0.159
#>
#> $Item_6
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.002 0.021 0.005 1 0.942
#> CSIBTEST group2 29 1 0.003 NA 0.029 2 0.986
#>
#> $Item_7
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.005 0.021 0.06 1 0.807
#> CSIBTEST group2 29 1 0.012 NA 0.342 2 0.843
#>
#> $Item_8
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.002 0.021 0.01 1 0.92
#> CSIBTEST group2 29 1 0.019 NA 1.123 2 0.57
#>
#> $Item_9
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.045 0.021 4.696 1 0.03
#> CSIBTEST group2 29 1 0.035 NA 4.82 2 0.09
#>
#> $Item_10
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.007 0.021 0.121 1 0.728
#> CSIBTEST group2 29 1 0.036 NA 3.296 2 0.192
#>
#> $Item_11
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.017 0.021 0.624 1 0.43
#> CSIBTEST group2 29 1 0.017 NA 0.624 1 0.43
#>
#> $Item_12
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.026 0.02 1.733 1 0.188
#> CSIBTEST group2 29 1 0.037 NA 3.391 2 0.183
#>
#> $Item_13
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.002 0.021 0.009 1 0.925
#> CSIBTEST group2 29 1 0.019 NA 1.139 2 0.566
#>
#> $Item_14
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.001 0.022 0.001 1 0.973
#> CSIBTEST group2 29 1 0.022 NA 1.108 2 0.575
#>
#> $Item_15
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.012 0.02 0.357 1 0.55
#> CSIBTEST group2 29 1 0.010 NA 0.358 2 0.836
#>
#> $Item_16
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.005 0.022 0.064 1 0.801
#> CSIBTEST group2 29 1 0.016 NA 0.547 2 0.761
#>
#> $Item_17
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.002 0.021 0.011 1 0.916
#> CSIBTEST group2 29 1 0.003 NA 0.025 2 0.987
#>
#> $Item_18
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.059 0.02 8.584 1 0.003
#> CSIBTEST group2 29 1 0.059 NA 8.584 1 0.003
#>
#> $Item_19
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.01 0.021 0.248 1 0.619
#> CSIBTEST group2 29 1 0.02 NA 1.031 2 0.597
#>
#> $Item_20
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.018 0.017 1.224 1 0.268
#> CSIBTEST group2 29 1 0.018 NA 1.224 1 0.268
#>
#> $Item_21
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.012 0.021 0.338 1 0.561
#> CSIBTEST group2 29 1 0.030 NA 2.06 2 0.357
#>
#> $Item_22
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.008 0.021 0.141 1 0.707
#> CSIBTEST group2 29 1 0.008 NA 0.171 2 0.918
#>
#> $Item_23
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.007 0.021 0.126 1 0.723
#> CSIBTEST group2 29 1 0.031 NA 2.224 2 0.329
#>
#> $Item_24
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.007 0.022 0.105 1 0.746
#> CSIBTEST group2 29 1 0.012 NA 0.578 2 0.749
#>
#> $Item_25
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.026 0.021 1.468 1 0.226
#> CSIBTEST group2 29 1 0.026 NA 1.468 1 0.226
#>
#> $Item_26
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.009 0.018 0.219 1 0.64
#> CSIBTEST group2 29 1 0.019 NA 3.558 2 0.169
#>
#> $Item_27
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.018 0.021 0.73 1 0.393
#> CSIBTEST group2 29 1 0.018 NA 0.73 1 0.393
#>
#> $Item_28
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.002 0.019 0.015 1 0.903
#> CSIBTEST group2 29 1 0.000 NA 0.017 2 0.991
#>
#> $Item_29
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.010 0.021 0.206 1 0.65
#> CSIBTEST group2 29 1 0.029 NA 1.883 2 0.39
#>
#> $Item_30
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.012 0.02 0.348 1 0.555
#> CSIBTEST group2 29 1 0.012 NA 0.402 2 0.818
#>
# post-hoc pairwise comparison for all groups
SIBTEST(dat, group, suspect_set = 1:ncol(dat), DIF=TRUE, pairwise = TRUE)
#> $group1_group2
#> $group1_group2$Item_1
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.023 0.02 1.399 1 0.237
#> CSIBTEST group2 29 1 0.027 NA 1.95 2 0.377
#>
#> $group1_group2$Item_2
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.045 0.021 4.628 1 0.031
#> CSIBTEST group2 29 1 0.041 NA 4.639 2 0.098
#>
#> $group1_group2$Item_3
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.003 0.021 0.027 1 0.87
#> CSIBTEST group2 29 1 0.003 NA 0.027 1 0.87
#>
#> $group1_group2$Item_4
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.002 0.015 0.018 1 0.894
#> CSIBTEST group2 29 1 0.015 NA 1.744 2 0.418
#>
#> $group1_group2$Item_5
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.03 0.021 1.979 1 0.159
#> CSIBTEST group2 29 1 0.03 NA 1.979 1 0.159
#>
#> $group1_group2$Item_6
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.002 0.021 0.005 1 0.942
#> CSIBTEST group2 29 1 0.003 NA 0.029 2 0.986
#>
#> $group1_group2$Item_7
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.005 0.021 0.06 1 0.807
#> CSIBTEST group2 29 1 0.012 NA 0.342 2 0.843
#>
#> $group1_group2$Item_8
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.002 0.021 0.01 1 0.92
#> CSIBTEST group2 29 1 0.019 NA 1.123 2 0.57
#>
#> $group1_group2$Item_9
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.045 0.021 4.696 1 0.03
#> CSIBTEST group2 29 1 0.035 NA 4.82 2 0.09
#>
#> $group1_group2$Item_10
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.007 0.021 0.121 1 0.728
#> CSIBTEST group2 29 1 0.036 NA 3.296 2 0.192
#>
#> $group1_group2$Item_11
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.017 0.021 0.624 1 0.43
#> CSIBTEST group2 29 1 0.017 NA 0.624 1 0.43
#>
#> $group1_group2$Item_12
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.026 0.02 1.733 1 0.188
#> CSIBTEST group2 29 1 0.037 NA 3.391 2 0.183
#>
#> $group1_group2$Item_13
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.002 0.021 0.009 1 0.925
#> CSIBTEST group2 29 1 0.019 NA 1.139 2 0.566
#>
#> $group1_group2$Item_14
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.001 0.022 0.001 1 0.973
#> CSIBTEST group2 29 1 0.022 NA 1.108 2 0.575
#>
#> $group1_group2$Item_15
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.012 0.02 0.357 1 0.55
#> CSIBTEST group2 29 1 0.010 NA 0.358 2 0.836
#>
#> $group1_group2$Item_16
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.005 0.022 0.064 1 0.801
#> CSIBTEST group2 29 1 0.016 NA 0.547 2 0.761
#>
#> $group1_group2$Item_17
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.002 0.021 0.011 1 0.916
#> CSIBTEST group2 29 1 0.003 NA 0.025 2 0.987
#>
#> $group1_group2$Item_18
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.059 0.02 8.584 1 0.003
#> CSIBTEST group2 29 1 0.059 NA 8.584 1 0.003
#>
#> $group1_group2$Item_19
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.01 0.021 0.248 1 0.619
#> CSIBTEST group2 29 1 0.02 NA 1.031 2 0.597
#>
#> $group1_group2$Item_20
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.018 0.017 1.224 1 0.268
#> CSIBTEST group2 29 1 0.018 NA 1.224 1 0.268
#>
#> $group1_group2$Item_21
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.012 0.021 0.338 1 0.561
#> CSIBTEST group2 29 1 0.030 NA 2.06 2 0.357
#>
#> $group1_group2$Item_22
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.008 0.021 0.141 1 0.707
#> CSIBTEST group2 29 1 0.008 NA 0.171 2 0.918
#>
#> $group1_group2$Item_23
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.007 0.021 0.126 1 0.723
#> CSIBTEST group2 29 1 0.031 NA 2.224 2 0.329
#>
#> $group1_group2$Item_24
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.007 0.022 0.105 1 0.746
#> CSIBTEST group2 29 1 0.012 NA 0.578 2 0.749
#>
#> $group1_group2$Item_25
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.026 0.021 1.468 1 0.226
#> CSIBTEST group2 29 1 0.026 NA 1.468 1 0.226
#>
#> $group1_group2$Item_26
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.009 0.018 0.219 1 0.64
#> CSIBTEST group2 29 1 0.019 NA 3.558 2 0.169
#>
#> $group1_group2$Item_27
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.018 0.021 0.73 1 0.393
#> CSIBTEST group2 29 1 0.018 NA 0.73 1 0.393
#>
#> $group1_group2$Item_28
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.002 0.019 0.015 1 0.903
#> CSIBTEST group2 29 1 0.000 NA 0.017 2 0.991
#>
#> $group1_group2$Item_29
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.010 0.021 0.206 1 0.65
#> CSIBTEST group2 29 1 0.029 NA 1.883 2 0.39
#>
#> $group1_group2$Item_30
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.012 0.02 0.348 1 0.555
#> CSIBTEST group2 29 1 0.012 NA 0.402 2 0.818
#>
#>
#> $group1_group3
#> $group1_group3$Item_1
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.002 0.02 0.006 1 0.937
#> CSIBTEST group3 29 1 0.032 NA 4.561 2 0.102
#>
#> $group1_group3$Item_2
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.022 0.021 1.079 1 0.299
#> CSIBTEST group3 29 1 0.024 NA 1.265 2 0.531
#>
#> $group1_group3$Item_3
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.003 0.02 0.021 1 0.886
#> CSIBTEST group3 29 1 0.009 NA 0.21 2 0.9
#>
#> $group1_group3$Item_4
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.003 0.016 0.043 1 0.836
#> CSIBTEST group3 29 1 0.004 NA 0.079 2 0.961
#>
#> $group1_group3$Item_5
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.019 0.021 0.757 1 0.384
#> CSIBTEST group3 29 1 0.033 NA 2.45 2 0.294
#>
#> $group1_group3$Item_6
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.006 0.021 0.079 1 0.779
#> CSIBTEST group3 29 1 0.017 NA 0.819 2 0.664
#>
#> $group1_group3$Item_7
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.016 0.021 0.589 1 0.443
#> CSIBTEST group3 29 1 0.016 NA 0.589 1 0.443
#>
#> $group1_group3$Item_8
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.007 0.021 0.103 1 0.748
#> CSIBTEST group3 29 1 0.010 NA 0.26 2 0.878
#>
#> $group1_group3$Item_9
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.002 0.021 0.013 1 0.911
#> CSIBTEST group3 29 1 0.032 NA 2.8 2 0.247
#>
#> $group1_group3$Item_10
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.002 0.02 0.01 1 0.919
#> CSIBTEST group3 29 1 0.015 NA 0.974 2 0.615
#>
#> $group1_group3$Item_11
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.014 0.021 0.447 1 0.504
#> CSIBTEST group3 29 1 0.014 NA 1.612 2 0.447
#>
#> $group1_group3$Item_12
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.024 0.02 1.397 1 0.237
#> CSIBTEST group3 29 1 0.023 NA 1.453 2 0.484
#>
#> $group1_group3$Item_13
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.006 0.021 0.092 1 0.762
#> CSIBTEST group3 29 1 0.000 NA 0.126 2 0.939
#>
#> $group1_group3$Item_14
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.004 0.021 0.032 1 0.859
#> CSIBTEST group3 29 1 0.032 NA 2.534 2 0.282
#>
#> $group1_group3$Item_15
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.023 0.02 1.367 1 0.242
#> CSIBTEST group3 29 1 0.040 NA 5.532 2 0.063
#>
#> $group1_group3$Item_16
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.034 0.021 2.489 1 0.115
#> CSIBTEST group3 29 1 0.034 NA 2.489 1 0.115
#>
#> $group1_group3$Item_17
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.000 0.021 0 1 0.985
#> CSIBTEST group3 29 1 0.007 NA 0.141 2 0.932
#>
#> $group1_group3$Item_18
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.022 0.02 1.216 1 0.27
#> CSIBTEST group3 29 1 0.001 NA 2.58 2 0.275
#>
#> $group1_group3$Item_19
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.025 0.02 1.525 1 0.217
#> CSIBTEST group3 29 1 0.010 NA 1.879 2 0.391
#>
#> $group1_group3$Item_20
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.003 0.015 0.034 1 0.854
#> CSIBTEST group3 29 1 0.014 NA 1.113 2 0.573
#>
#> $group1_group3$Item_21
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.031 0.021 2.216 1 0.137
#> CSIBTEST group3 29 1 0.031 NA 2.216 1 0.137
#>
#> $group1_group3$Item_22
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.018 0.021 0.676 1 0.411
#> CSIBTEST group3 29 1 0.018 NA 0.676 1 0.411
#>
#> $group1_group3$Item_23
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.019 0.021 0.823 1 0.364
#> CSIBTEST group3 29 1 0.019 NA 0.823 1 0.364
#>
#> $group1_group3$Item_24
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.018 0.021 0.75 1 0.386
#> CSIBTEST group3 29 1 0.026 NA 1.727 2 0.422
#>
#> $group1_group3$Item_25
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.013 0.021 0.392 1 0.532
#> CSIBTEST group3 29 1 0.018 NA 0.746 2 0.689
#>
#> $group1_group3$Item_26
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.010 0.018 0.304 1 0.581
#> CSIBTEST group3 29 1 0.005 NA 0.325 2 0.85
#>
#> $group1_group3$Item_27
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.017 0.021 0.649 1 0.421
#> CSIBTEST group3 29 1 0.025 NA 1.722 2 0.423
#>
#> $group1_group3$Item_28
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.001 0.02 0.003 1 0.954
#> CSIBTEST group3 29 1 0.009 NA 0.349 2 0.84
#>
#> $group1_group3$Item_29
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.028 0.021 1.764 1 0.184
#> CSIBTEST group3 29 1 0.028 NA 1.764 1 0.184
#>
#> $group1_group3$Item_30
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.011 0.02 0.316 1 0.574
#> CSIBTEST group3 29 1 0.025 NA 1.705 2 0.426
#>
#>
#> $group2_group3
#> $group2_group3$Item_1
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.020 0.019 1.056 1 0.304
#> CSIBTEST group3 29 1 0.003 NA 1.628 2 0.443
#>
#> $group2_group3$Item_2
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.023 0.021 1.233 1 0.267
#> CSIBTEST group3 29 1 0.023 NA 1.233 1 0.267
#>
#> $group2_group3$Item_3
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.000 0.02 0 1 0.993
#> CSIBTEST group3 29 1 0.003 NA 0.122 2 0.941
#>
#> $group2_group3$Item_4
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.005 0.015 0.12 1 0.729
#> CSIBTEST group3 29 1 0.001 NA 0.339 2 0.844
#>
#> $group2_group3$Item_5
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.01 0.021 0.23 1 0.631
#> CSIBTEST group3 29 1 0.00 NA 0.241 2 0.886
#>
#> $group2_group3$Item_6
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.006 0.021 0.078 1 0.779
#> CSIBTEST group3 29 1 0.006 NA 0.078 1 0.779
#>
#> $group2_group3$Item_7
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.019 0.021 0.791 1 0.374
#> CSIBTEST group3 29 1 0.019 NA 0.791 1 0.374
#>
#> $group2_group3$Item_8
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.007 0.021 0.102 1 0.75
#> CSIBTEST group3 29 1 0.013 NA 0.84 2 0.657
#>
#> $group2_group3$Item_9
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.045 0.021 4.764 1 0.029
#> CSIBTEST group3 29 1 0.045 NA 4.764 1 0.029
#>
#> $group2_group3$Item_10
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.008 0.021 0.139 1 0.709
#> CSIBTEST group3 29 1 0.014 NA 0.468 2 0.791
#>
#> $group2_group3$Item_11
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.001 0.021 0.001 1 0.978
#> CSIBTEST group3 29 1 0.017 NA 0.866 2 0.648
#>
#> $group2_group3$Item_12
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.000 0.02 0 1 0.991
#> CSIBTEST group3 29 1 0.002 NA 0.007 2 0.996
#>
#> $group2_group3$Item_13
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.002 0.021 0.014 1 0.907
#> CSIBTEST group3 29 1 0.008 NA 0.204 2 0.903
#>
#> $group2_group3$Item_14
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.005 0.021 0.051 1 0.822
#> CSIBTEST group3 29 1 0.011 NA 1.915 2 0.384
#>
#> $group2_group3$Item_15
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.011 0.02 0.335 1 0.563
#> CSIBTEST group3 29 1 0.049 NA 6.682 2 0.035
#>
#> $group2_group3$Item_16
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.031 0.021 2.131 1 0.144
#> CSIBTEST group3 29 1 0.029 NA 2.194 2 0.334
#>
#> $group2_group3$Item_17
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.004 0.021 0.029 1 0.864
#> CSIBTEST group3 29 1 0.001 NA 0.03 2 0.985
#>
#> $group2_group3$Item_18
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.038 0.021 3.351 1 0.067
#> CSIBTEST group3 29 1 0.038 NA 3.351 1 0.067
#>
#> $group2_group3$Item_19
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.021 0.021 1.06 1 0.303
#> CSIBTEST group3 29 1 0.006 NA 3.523 2 0.172
#>
#> $group2_group3$Item_20
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.017 0.017 0.992 1 0.319
#> CSIBTEST group3 29 1 0.017 NA 0.992 1 0.319
#>
#> $group2_group3$Item_21
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.016 0.021 0.578 1 0.447
#> CSIBTEST group3 29 1 0.034 NA 4.188 2 0.123
#>
#> $group2_group3$Item_22
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.033 0.021 2.372 1 0.124
#> CSIBTEST group3 29 1 0.033 NA 2.372 1 0.124
#>
#> $group2_group3$Item_23
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.009 0.021 0.178 1 0.673
#> CSIBTEST group3 29 1 0.024 NA 1.949 2 0.377
#>
#> $group2_group3$Item_24
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.008 0.022 0.151 1 0.698
#> CSIBTEST group3 29 1 0.018 NA 0.683 2 0.711
#>
#> $group2_group3$Item_25
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.043 0.02 4.495 1 0.034
#> CSIBTEST group3 29 1 0.043 NA 4.495 1 0.034
#>
#> $group2_group3$Item_26
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.003 0.019 0.025 1 0.875
#> CSIBTEST group3 29 1 0.023 NA 3.11 2 0.211
#>
#> $group2_group3$Item_27
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.002 0.021 0.007 1 0.933
#> CSIBTEST group3 29 1 0.013 NA 0.373 2 0.83
#>
#> $group2_group3$Item_28
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.002 0.02 0.013 1 0.909
#> CSIBTEST group3 29 1 0.008 NA 0.354 2 0.838
#>
#> $group2_group3$Item_29
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.018 0.022 0.722 1 0.395
#> CSIBTEST group3 29 1 0.026 NA 1.466 2 0.481
#>
#> $group2_group3$Item_30
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.029 0.02 1.973 1 0.16
#> CSIBTEST group3 29 1 0.032 NA 2.615 2 0.271
#>
#>
## systematic differing slopes and intercepts
dat2 <- simdata(a + c(numeric(15), .5,.5,.5,.5,.5, numeric(10)),
d + c(numeric(15), 0,.6,.7,.8,.9, numeric(10)),
N, itemtype = 'dich')
dat <- rbind(dat1, dat2, dat3)
SIBTEST(dat, group, suspect_set = 16)
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.008 -0.034 4.642 2 0.098
#> GCSIBTEST 29 1 0.059 0.034 8.205
SIBTEST(dat, group, suspect_set = 16, randomize=TRUE)
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.008 -0.034 4.642 2 0.098
#> GCSIBTEST 29 1 0.059 0.034 8.205 0.125
SIBTEST(dat, group, suspect_set = 19)
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.136 -0.025 52.967 2 0
#> GCSIBTEST 29 1 0.130 0.009 61.401
SIBTEST(dat, group, suspect_set = 19, randomize=TRUE)
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.136 -0.025 52.967 2 0
#> GCSIBTEST 29 1 0.130 0.009 61.401 0
SIBTEST(dat, group, suspect_set = c(16, 19), DIF=TRUE)
#> $Item_16
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 0.008 -0.034 4.642 2 0.098
#> GCSIBTEST 29 1 0.059 0.034 8.205
#>
#> $Item_19
#> n_matched_set n_suspect_set beta_1 beta_2 X2 df p
#> GSIBTEST 29 1 -0.136 -0.025 52.967 2 0
#> GCSIBTEST 29 1 0.130 0.009 61.401
#>
SIBTEST(dat, group, suspect_set = c(16, 19), DIF=TRUE, pairwise=TRUE)
#> $group1_group2
#> $group1_group2$Item_16
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 0.008 0.021 0.154 1 0.695
#> CSIBTEST group2 29 1 0.057 NA 9.536 2 0.008
#>
#> $group1_group2$Item_19
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group2 29 1 -0.138 0.021 45.595 1 0
#> CSIBTEST group2 29 1 0.141 NA 47.776 2 0
#>
#>
#> $group1_group3
#> $group1_group3$Item_16
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.034 0.021 2.489 1 0.115
#> CSIBTEST group3 29 1 0.034 NA 2.489 1 0.115
#>
#> $group1_group3$Item_19
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.025 0.02 1.525 1 0.217
#> CSIBTEST group3 29 1 0.010 NA 1.879 2 0.391
#>
#>
#> $group2_group3
#> $group2_group3$Item_16
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 -0.039 0.02 3.721 1 0.054
#> CSIBTEST group3 29 1 0.040 NA 4.185 2 0.123
#>
#> $group2_group3$Item_19
#> focal_group n_matched_set n_suspect_set beta SE X2 df p
#> SIBTEST group3 29 1 0.112 0.02 30.503 1 0
#> CSIBTEST group3 29 1 0.118 NA 37.348 2 0
#>
#>
# }