G*Power examples evaluated with Spower
Phil Chalmers
2026-02-13
Source:vignettes/gpower_examples.Rmd
gpower_examples.RmdThis vignette replicates several of the examples found in the G*Power
manual (version 3.1). It is not meant to be exhaustive, but instead
demonstrates how the presented power analyses can be computed and
extended using simulation methodology by either editing the default
functions found within the package, or by creating a new user-defined
function for yet-to-be-defined statistical analysis contexts. Unless
otherwise specified, the following analyses assume that the
“significance level” (sig.level in Spower())
is set to
.
Correlation
Correlation analyses require evaluating the power associated with the hypotheses
where is the population correlation and the null hypothesis constant.
Example 3.3; Difference from constant (one sample case)
The following estimates the sample size required to reject in correlation analysis with probability when .
##
## Execution time (H:M:S): 00:00:22
## Design conditions:
##
## # A tibble: 1 × 5
## n r rho sig.level power
## <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 NA 0.65 0.6 0.05 0.95
##
## Estimate of n: 1931.4
## 95% Predicted Confidence Interval: [1901.1, 1958.2]
# this is equivalent:
# p_r(n = NA, r = .65, rho = .60) |>
# Spower(power = .95, interval=c(500,3000))G*power estimates this
to be 1929 using the Fisher
-transformation
approximation, which is what is used by the Spower
definition as well.
Test against constant
The more canonical version hypotheses involving correlation coefficients appear when , as these do not require the Fisher approximation. For instance, the power associated with with 100 pairs of observations, tested against , results in the following.
##
## Execution time (H:M:S): 00:00:11
## Design conditions:
##
## # A tibble: 1 × 4
## n r sig.level power
## <dbl> <dbl> <dbl> <lgl>
## 1 100 0.3 0.05 NA
##
## Estimate of power: 0.861
## 95% Confidence Interval: [0.854, 0.867]Next, the sample sample size estimate required to reject in correlation analysis with probability when is expressed as
##
## Execution time (H:M:S): 00:00:17
## Design conditions:
##
## # A tibble: 1 × 4
## n r sig.level power
## <dbl> <dbl> <dbl> <dbl>
## 1 NA 0.3 0.05 0.95
##
## Estimate of n: 138.3
## 95% Predicted Confidence Interval: [136.1, 140.4]G*power 3.1 provides the same estimate as the pwr
package in this case, which for comparison is presented below.
pwr::pwr.r.test(r=.3, power=.95, n=NULL)##
## approximate correlation power calculation (arctangh transformation)
##
## n = 137.8
## r = 0.3
## sig.level = 0.05
## power = 0.95
## alternative = two.sidedExample 27.3; Correlation - inequality of two independent Pearson r’s
Were the correlation between two independent samples to be compared,
the p_2r() simulation can be adopted. Below a sample of
observations appeared in the first sample
(),
while the second sample
()
contained only
observations (hence, the ratio
).
This results in the post-hoc/observed power of
##
## Execution time (H:M:S): 00:00:27
## Design conditions:
##
## # A tibble: 1 × 5
## n r.ab r.ab2 sig.level power
## <dbl> <dbl> <dbl> <dbl> <lgl>
## 1 206 0.75 0.88 0.05 NA
##
## Estimate of power: 0.727
## 95% Confidence Interval: [0.718, 0.735]G*power 3.1 returns the power of .726 in this context.
Example 28.3.1; Correlation - inequality of two dependent Pearson r’s (no common index)
The following two examples assume the correlation matrix
# From Gpower 3.1 manual
Cp <- matrix(c(1, .5, .4, .1,
.5, 1, .2, -.4,
.4, .2, 1, .8,
.1, -.4, .8, 1), 4, 4)
# rearrange rows for convenience
Cp <- Cp[c(1,4,2,3), c(1,4,2,3)]
colnames(Cp) <- rownames(Cp) <- c('x1', 'y1', 'x2', 'y2')
Cp## x1 y1 x2 y2
## x1 1.0 0.1 0.5 0.4
## y1 0.1 1.0 -0.4 0.8
## x2 0.5 -0.4 1.0 0.2
## y2 0.4 0.8 0.2 1.0is the population structure. For the no common index tests all of these elements are required, while for the common index form only a subset is needed.
Evaluating the null hypothesis that
where in this case
and
can be explored using the p_2r() function. The following
performs an a priori analyses to determine the sample size
()
required to achieve 80% power using Steiger’s (1980) inferential
approach.
p_2r(n=interval(500, 2000), r.ab=.1, r.ac=.5, r.ad=.4, r.bc=-.4, r.bd=.8, r.cd=.2, two.tailed=FALSE) |>
Spower(power = .80)##
## Execution time (H:M:S): 00:01:36
## Design conditions:
##
## # A tibble: 1 × 9
## n r.ab r.ac r.ad r.bd r.cd two.tailed sig.level power
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <lgl> <dbl> <dbl>
## 1 NA 0.1 0.5 0.4 0.8 0.2 FALSE 0.05 0.8
##
## Estimate of n: 886.2
## 95% Predicted Confidence Interval: [875.3, 897.3]G*power 3.1 returns the required sample size of .
Example 28.3.2; Correlation - inequality of two dependent Pearson r’s (common index)
The information in this example is the same as
Example 28.3.1, however it is assumed that there is a
common index between the correlation measures instead of a complete
overlap. As such, the previous Cp object may be further
subset to see what type of correlation structure is required for the
common index setup.
## y2 x2 x1
## y2 1.0 0.2 0.4
## x2 0.2 1.0 0.5
## x1 0.4 0.5 1.0The null under instigation in this case is
where
and
.
In Spower, this equates to the following inputs, which
again use Steiger’s (1980) inferential
approach by default.
##
## Execution time (H:M:S): 00:01:19
## Design conditions:
##
## # A tibble: 1 × 7
## n r.ab r.ac r.bc two.tailed sig.level power
## <dbl> <dbl> <dbl> <dbl> <lgl> <dbl> <dbl>
## 1 NA 0.4 0.2 0.5 FALSE 0.05 0.8
##
## Estimate of n: 134.7
## 95% Predicted Confidence Interval: [133.1, 136.3]G*power 3.1 returns the required sample size of
,
which interestingly is slightly higher than the simulation version from
Spower. Providing
to the above to obtain the power estimate gives the following:
##
## Execution time (H:M:S): 00:00:22
## Design conditions:
##
## # A tibble: 1 × 7
## n r.ab r.ac r.bc two.tailed sig.level power
## <dbl> <dbl> <dbl> <dbl> <lgl> <dbl> <lgl>
## 1 144 0.4 0.2 0.5 FALSE 0.05 NA
##
## Estimate of power: 0.831
## 95% Confidence Interval: [0.824, 0.839]Example 28.3.3; sensitivity analysis
It is also possible to perform a sensitivity analyses rather than the
above a priori power analysis. Below fixes
,
while r.ac is solved to obtain 80% power. G*power 3.1
reports that
,
which is confirmed using the simulation below.
# confirm solution obtained by G*power (post hoc power estimate)
p_2r(n=144, r.ab=.4, r.ac=0.047702, r.bc=-0.6, two.tailed=FALSE) |> Spower()##
## Execution time (H:M:S): 00:00:22
## Design conditions:
##
## # A tibble: 1 × 6
## n r.ab r.ac two.tailed sig.level power
## <dbl> <dbl> <dbl> <lgl> <dbl> <lgl>
## 1 144 0.4 0.047702 FALSE 0.05 NA
##
## Estimate of power: 0.815
## 95% Confidence Interval: [0.808, 0.823]Obtaining a similar estimate using Spower() requires the
following sensitivity analysis structure:
# note that interval is specified as c(upper, lower) as higher values
# of r.ac result in lower power in this context
p_2r(n=144, r.ab=.4, r.ac=interval(.4, .001), r.bc=-0.6, two.tailed=FALSE) |>
Spower(power = .80)##
## Execution time (H:M:S): 00:01:38
## Design conditions:
##
## # A tibble: 1 × 6
## n r.ab r.ac two.tailed sig.level power
## <dbl> <dbl> <dbl> <lgl> <dbl> <dbl>
## 1 144 0.4 NA FALSE 0.05 0.8
##
## Estimate of r.ac: 0.048
## 95% Predicted Confidence Interval: [0.046, 0.050]For this example, Spower and G*power 3.1 seem to
agree.
Example 16.3; Point-biserial correlation
The following estimates the sample size required to obtain a power of given that is the true correlation, evaluated under the null (hence, is one-tailed) with .
# solution per group
out <- p_t.test(r = .25, n = interval(50, 200), two.tailed=FALSE) |>
Spower(power = .95)
out##
## Execution time (H:M:S): 00:00:11
## Design conditions:
##
## # A tibble: 1 × 5
## n r two.tailed sig.level power
## <dbl> <dbl> <lgl> <dbl> <dbl>
## 1 NA 0.25 FALSE 0.05 0.95
##
## Estimate of n: 81.7
## 95% Predicted Confidence Interval: [79.1, 85.4]
# total sample size required
ceiling(out$n) * 2## [1] 164G*power gives the result .
Relatedly, one can specify
,
Cohen’s standardized mean-difference effect size, instead of
since
will be converted to
inside the p_t.test() function.
Example 31.3; tetrachoric correlation
For tetrachoric and polychoric correlations, the experiment
definition in p_r.cat() can be used. This requires
specifying the associated
threshold coefficients for the population normal truncation processes,
as well as the bivariate correlation itself prior to the truncation.
## [1] 0.4183 0.5817
(marginal.y <- rowSums(F)/N)## [1] 0.3978 0.6022
# converted to intercepts
tauX <- qnorm(1-marginal.x)[2]
tauY <- qnorm(1-marginal.y)[2]
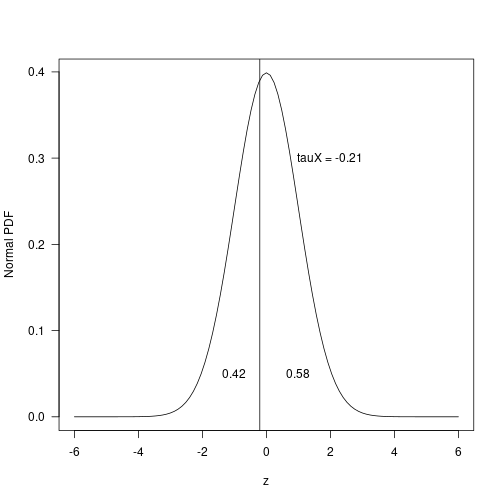
c(tauX, tauY)## [1] -0.2063 -0.2589These values correspond to where along the assumed normal p.d.f. the truncation took place, which for the variable can be seen in the following graphic.
Finally, assuming that the untruncated
,
and a Score test were used to evaluate the null hypothesis of interest
(score = TRUE), the sample size required to reject the null
hypothesis that the tetrachoric correlation is less than or equal to 0
in this population (one-tailed) is expressed as
p_r.cat(n=interval(100, 500), r=0.2399846, tauX=tauX, tauY=tauY,
score=TRUE, two.tailed=FALSE) |>
Spower(power = .95, parallel=TRUE)##
## Design conditions:
##
## # A tibble: 1 × 8
## n r tauX tauY score two.tailed sig.level power
## <dbl> <dbl> <dbl> <dbl> <lgl> <lgl> <dbl> <dbl>
## 1 NA 0.240 -0.206 -0.259 FALSE FALSE 0.05 0.95
##
## Estimate of n: 462.9
## 95% Prediction Interval: [458.5, 466.6]G*power gives
,
though uses the SE value at the null (Score test).
p_r.cat(), on the other hand, defaults to the Wald approach
where the SE is at the maximum-likelihood estimate (MLE); hence,
score = FALSE by default. To switch, use
score=TRUE, though note that this requires twice as many
computations as a second set of data is generated and analyzed at
to obtain the required
estimate.
Proportions
Example 4.3; One sample proportion tests
A one sample, one-tailed proportion test given data generated from a
population with
and tested against the null hypothesis
with
is presented in the following. Note that G*power requires a term
to be specified as the proportion difference from the null
instead (hence,
),
though p_prop.teset() accepts the null and alternative
probability values as-is.
pi <- .65
g <- .15
p <- pi + g
p_prop.test(n=20, prop=p, pi=pi, two.tailed=FALSE) |>
Spower()##
## Execution time (H:M:S): 00:00:04
## Design conditions:
##
## # A tibble: 1 × 4
## n two.tailed sig.level power
## <dbl> <lgl> <dbl> <lgl>
## 1 20 FALSE 0.05 NA
##
## Estimate of power: 0.416
## 95% Confidence Interval: [0.406, 0.425]G*power gives the estimate
.
Note that with p_prop.test(), the Fisher’s exact version of
this test is also supported by passing the argument
exact = TRUE.
# Fisher exact test
p_prop.test(n=20, prop=p, pi=pi, exact=TRUE,
two.tailed=FALSE) |> Spower()##
## Execution time (H:M:S): 00:00:04
## Design conditions:
##
## # A tibble: 1 × 5
## n two.tailed exact sig.level power
## <dbl> <lgl> <lgl> <dbl> <lgl>
## 1 20 FALSE TRUE 0.05 NA
##
## Estimate of power: 0.411
## 95% Confidence Interval: [0.402, 0.421]Example 22.1; Wilcoxon signed-rank test
The following performed a one-sample, one-tailed Wilcoxon signed rank test given , , where the parent distribution is assumed to follow a Normal/Gaussian shape (default).
p_wilcox.test(n=649, d=.1, type='one.sample', two.tailed=FALSE) |>
Spower()##
## Execution time (H:M:S): 00:00:21
## Design conditions:
##
## # A tibble: 1 × 6
## n d type two.tailed sig.level power
## <dbl> <dbl> <chr> <lgl> <dbl> <lgl>
## 1 649 0.1 one.sample FALSE 0.05 NA
##
## Estimate of power: 0.812
## 95% Confidence Interval: [0.805, 0.820]G*power gives the power estimate of .800.
The following partially recreates the simulation results in Figure 29
(which itself was partially extracted from Shieh, Jan, and Randles,
2007) for the
Gaussian(,1)
distribution with varying sample sizes and effect sizes. The target was
to obtain the “approximate power of
”,
though how these sample sizes were decided upon was not specified.
Spower()’s stochastic root-solving approach would likely
get closer to more optimal
estimates were these the target of the analyses.
# For Gaussian(d,1)
out <- p_wilcox.test(type='one.sample', two.tailed=FALSE) |>
SpowerBatch(n=c(649, 164, 42, 20, 12, 9),
d=c(.1, .2, .4, .6, .8, 1.0), replications = 50000, fully.crossed=FALSE)
as.data.frame(out)## n d type two.tailed sig.level power CI_2.5 CI_97.5
## 1 649 0.1 one.sample FALSE 0.05 0.8012 0.7977 0.8047
## 2 164 0.2 one.sample FALSE 0.05 0.8027 0.7992 0.8062
## 3 42 0.4 one.sample FALSE 0.05 0.8043 0.8009 0.8078
## 4 20 0.6 one.sample FALSE 0.05 0.8070 0.8035 0.8105
## 5 12 0.8 one.sample FALSE 0.05 0.8018 0.7983 0.8053
## 6 9 1.0 one.sample FALSE 0.05 0.8467 0.8435 0.8498Laplace(, 1) version
A one-sample Wilcoxon signed rank test with Laplace distribution as
the parent. Note that this requires defining the parent distribution
manually, accepting arguments such as n and
d.
library(extraDistr)
# generate data with scale 0-1 for d effect size to be same as mean
# VAR = 2*b^2, so scale should be 1 = 2*b^2 -> sqrt(1/2)
parent <- function(n, d, sigma=sqrt(1/2))
extraDistr::rlaplace(n, d, sigma=sigma)
p_wilcox.test(n=11, d=.8, parent1=parent, type='one.sample',
two.tailed=FALSE, correct = FALSE) |> Spower()##
## Execution time (H:M:S): 00:00:04
## Design conditions:
##
## # A tibble: 1 × 7
## n d type correct two.tailed sig.level power
## <dbl> <dbl> <chr> <lgl> <lgl> <dbl> <lgl>
## 1 11 0.8 one.sample FALSE FALSE 0.05 NA
##
## Estimate of power: 0.801
## 95% Confidence Interval: [0.793, 0.809]G*power gives the estimate .830, which seems somewhat high (see below).
The following partially recreates the simulation results in Figure 29
for the
Laplace(,
1) distribution with varying sample sizes and effect sizes. The target
was to obtain “approximate power of
”,
though how these sample sizes were decided upon was not specified.
Spower()’s stochastic root-solving approach would likely
get closer to more optimal
estimates were these the target of the analyses.
# For Laplace(0,1)
out <- p_wilcox.test(parent1=parent, type='one.sample',
two.tailed=FALSE) |>
SpowerBatch(n=c(419, 109, 31, 16, 11, 8),
d=c(.1, .2, .4, .6, .8, 1.0), replications=50000, fully.crossed=FALSE)
as.data.frame(out)## n d type two.tailed sig.level power CI_2.5 CI_97.5
## 1 419 0.1 one.sample FALSE 0.05 0.8021 0.7986 0.8056
## 2 109 0.2 one.sample FALSE 0.05 0.7992 0.7957 0.8028
## 3 31 0.4 one.sample FALSE 0.05 0.8031 0.7996 0.8065
## 4 16 0.6 one.sample FALSE 0.05 0.8007 0.7972 0.8042
## 5 11 0.8 one.sample FALSE 0.05 0.8032 0.7998 0.8067
## 6 8 1.0 one.sample FALSE 0.05 0.7771 0.7735 0.7808Example 5.3; Two dependent proportions test (McNemar’s test)
The following performs a proportions test between two dependent groups using McNemar’s test. The data is from O’Brien (2002, p. 161-163).
obrien2002 <- matrix(c(.54, .32, .08, .06), 2, 2,
dimnames = list('Treatment' = c('Yes', 'No'),
'Standard' = c('Yes', 'No')))
obrien2002## Standard
## Treatment Yes No
## Yes 0.54 0.08
## No 0.32 0.06
p_mcnemar.test(n=50, prop=obrien2002, two.tailed=FALSE) |> Spower()##
## Execution time (H:M:S): 00:00:03
## Design conditions:
##
## # A tibble: 1 × 4
## n two.tailed sig.level power
## <dbl> <lgl> <dbl> <lgl>
## 1 50 FALSE 0.05 NA
##
## Estimate of power: 0.836
## 95% Confidence Interval: [0.828, 0.843]Alternatively, specifying the inputs not in terms of proportions but rather as the odds ratio () and proportions of discordant pairs () can be supplied
OR <- obrien2002[1,2] / obrien2002[2,1]
disc <- obrien2002[1,2] + obrien2002[2,1]
p_mcnemar.test(n=50, OR=OR, prop.disc=disc, two.tailed=FALSE) |> Spower()##
## Execution time (H:M:S): 00:00:03
## Design conditions:
##
## # A tibble: 1 × 4
## n two.tailed sig.level power
## <dbl> <lgl> <dbl> <lgl>
## 1 50 FALSE 0.05 NA
##
## Estimate of power: 0.841
## 95% Confidence Interval: [0.834, 0.848]G*Power gives .839 (). Slightly more power can be achieved when not using the continuity correction, though in general this is not recommended in practice.
p_mcnemar.test(n=50, prop=obrien2002, two.tailed=FALSE, correct=FALSE) |> Spower()##
## Execution time (H:M:S): 00:00:03
## Design conditions:
##
## # A tibble: 1 × 5
## n two.tailed correct sig.level power
## <dbl> <lgl> <lgl> <dbl> <lgl>
## 1 50 FALSE FALSE 0.05 NA
##
## Estimate of power: 0.887
## 95% Confidence Interval: [0.881, 0.893]Multiple Linear Regression (Fixed IVs)
Example 13.1
Evaluating generated data for a linear regression model given the null hypothesis . When evaluated using observations with predictor variables gives the estimate.
##
## Execution time (H:M:S): 00:00:44
## Design conditions:
##
## # A tibble: 1 × 5
## n R2 k sig.level power
## <dbl> <dbl> <dbl> <dbl> <lgl>
## 1 95 0.1 5 0.05 NA
##
## Estimate of power: 0.664
## 95% Confidence Interval: [0.655, 0.674]G*power gives .
Example 14.3
Similarly, comparing nested models for changes in
.
For the following, note that k is total IVs (in this case,
9), while k.R2_0 is number of IVs for baseline model (in
this case, 5). At
and a change of
from the baseline
gives
##
## Execution time (H:M:S): 00:00:56
## Design conditions:
##
## # A tibble: 1 × 7
## n R2 k R2_0 k.R2_0 sig.level power
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <lgl>
## 1 90 0.3 9 0.25 5 0.01 NA
##
## Estimate of power: 0.238
## 95% Confidence Interval: [0.230, 0.247]G*power gives . Solving the sample size to achieve 80% power
##
## Execution time (H:M:S): 00:03:09
## Design conditions:
##
## # A tibble: 1 × 7
## n R2 k R2_0 k.R2_0 sig.level power
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 NA 0.3 9 0.25 5 0.01 0.8
##
## Estimate of n: 242.6
## 95% Predicted Confidence Interval: [240.5, 244.6]G*power gives .
Example 14.3b
Nested model comparison for changes in for models with 12 IVs versus 9 IVs. Requires the specification of the .
##
## Execution time (H:M:S): 00:01:17
## Design conditions:
##
## # A tibble: 1 × 8
## n R2 k R2_0 k.R2_0 R2.resid sig.level power
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <lgl>
## 1 200 0.16 12 0.1 9 0.8 0.01 NA
##
## Estimate of power: 0.756
## 95% Confidence Interval: [0.748, 0.765]G*power gives .
Multiple Linear Regression (Random IVs)
Example 7.3
Same as in Example 13.1 above, however assuming that the IVs are randomly sampled instead of fixed.
##
## Execution time (H:M:S): 00:00:16
## Design conditions:
##
## # A tibble: 1 × 6
## n R2 k fixed sig.level power
## <dbl> <dbl> <dbl> <lgl> <dbl> <lgl>
## 1 95 0.1 5 FALSE 0.05 NA
##
## Estimate of power: 0.659
## 95% Confidence Interval: [0.650, 0.669]G*power gives 0.662 using a one-tailed test criterion.
Simple linear regression
Example 12.3
Evaluate post-hoc power for simple linear regression model null hypothesis given , , , and .
##
## Execution time (H:M:S): 00:00:31
## Design conditions:
##
## # A tibble: 1 × 5
## n sd_x sd_y sig.level power
## <dbl> <dbl> <dbl> <dbl> <lgl>
## 1 100 7.5 4 0.05 NA
##
## Estimate of power: 0.243
## 95% Confidence Interval: [0.234, 0.251]G*power returns the power estimate .
Fixed effects ANOVA - One way (F-test)
Example 10.3
One-way ANOVA example to solve per group (of which there are ), using Cohen’s , to achieve a power of .
p_anova.test(n=interval(20, 300), k=10, f=.25) |> Spower(power=.95)##
## Execution time (H:M:S): 00:00:23
## Design conditions:
##
## # A tibble: 1 × 5
## n k f sig.level power
## <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 NA 10 0.25 0.05 0.95
##
## Estimate of n: 38.7
## 95% Predicted Confidence Interval: [38.1, 39.2]G*power gives the estimate .
Fixing in total (hence, ) and performing a compromise analysis assuming ,
p_anova.test(n=20, k=10, f=.25) |> Spower(beta_alpha=1, replications=30000)##
## Execution time (H:M:S): 00:00:44
## Design conditions:
##
## # A tibble: 1 × 6
## n k f sig.level power beta_alpha
## <dbl> <dbl> <dbl> <dbl> <lgl> <dbl>
## 1 20 10 0.25 NA NA 1
##
## Estimate of Type I error rate (alpha/sig.level): 0.160
## 95% Confidence Interval: [0.156, 0.164]
##
## Estimate of power (1-beta): 0.840
## 95% Confidence Interval: [0.836, 0.844]G*Power gives .
-test: Linear regression (two groups)
Test coefficients across distinct datasets with similar form. In this case
where the null of interest is
To do this a multiple linear regression model is setup with three variables
where , , and is a binary indicator variable indicating whether the observations were in the second sample.
When the first group’s parameterization will be recovered, while when the second group’s parameterization will be recovered as the potentially non-zero reflects a change in the intercept () while the change in the slope for the second group will be reflected by the (). Hence, the null hypothesis that the two groups have the same slope can be evaluated using this augmented model by testing
Example 17.3 and 18.3
We start by defining the population generating model to replace the
gen_glm() function that is the default in
p_glm(). This generating function is organized such that a
data.frame is returned with the columns y,
X, and S, where the interaction effect
reflects the magnitude of the difference between the
coefficients across the independent samples.
gen_twogroup <- function(n, dbeta, sdx1, sdx2, sigma, n2_n1 = 1, ...){
X1 <- rnorm(n, sd=sdx1)
X2 <- rnorm(n*n2_n1, sd=sdx2)
X <- c(X1, X2)
N <- length(X)
S <- c(rep(0, n), rep(1, N-n))
y <- dbeta * X*S + rnorm(N, sd=sigma)
dat <- data.frame(y, X, S)
dat
}To demonstrate, the post-hoc power for the described example in G*Power is the following.
p_glm(formula=y~X*S, test="X:S = 0",
n=28, n2_n1=44/28, sdx1=9.02914, sdx2=11.86779, dbeta=0.01592,
sigma=0.5578413, gen_fun=gen_twogroup) |> Spower()##
## Execution time (H:M:S): 00:00:35
## Design conditions:
##
## # A tibble: 1 × 8
## test sigma n sdx1 sdx2 dbeta sig.level power
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <lgl>
## 1 X:S = 0 0.55784 28 9.0291 11.868 0.01592 0.05 NA
##
## Estimate of power: 0.199
## 95% Confidence Interval: [0.191, 0.207]For the a priori power analysis to achieve a power of .80
p_glm(formula=y~X*S, test="X:S = 0",
n=interval(100, 1000), n2_n1=44/28, sdx1=9.02914, sdx2=11.86779, dbeta=0.01592,
sigma=0.5578413, gen_fun=gen_twogroup) |>
Spower(power=.8)##
## Execution time (H:M:S): 00:02:01
## Design conditions:
##
## # A tibble: 1 × 8
## test sigma n sdx1 sdx2 dbeta sig.level power
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 X:S = 0 0.55784 NA 9.0291 11.868 0.01592 0.05 0.8
##
## Estimate of n: 164.9
## 95% Predicted Confidence Interval: [163.3, 166.8]G*Power gives the estimate for
to be 163 (and therefore 256 in the second group given the
n2_n1).
Variance tests
Example 26.3; Difference from constant (one sample case)
Solve for variance ratio of using a one-tailed variance ratio test, assuming that the target power is .
p_var.test(n=interval(10, 200), vars=1, sigma2=1.5, two.tailed=FALSE) |>
Spower(power=.80)##
## Execution time (H:M:S): 00:00:34
## Design conditions:
##
## # A tibble: 1 × 6
## n vars sigma2 two.tailed sig.level power
## <dbl> <dbl> <dbl> <lgl> <dbl> <dbl>
## 1 NA 1 1.5 FALSE 0.05 0.8
##
## Estimate of n: 80.6
## 95% Predicted Confidence Interval: [79.3, 82.2]G*power gives sample size of 81.
Example 15.3; Two-sample variance test
For a two-sample equality of variance test with equal sample sizes,
# solve n for variance ratio of 1/1.5 = 2/3, two.tailed, 80% power
p_var.test(n=interval(50, 300), vars=c(1, 1.5), two.tailed=TRUE) |>
Spower(power=.80)##
## Execution time (H:M:S): 00:00:46
## Design conditions:
##
## # A tibble: 1 × 4
## n two.tailed sig.level power
## <dbl> <lgl> <dbl> <dbl>
## 1 NA TRUE 0.05 0.8
##
## Estimate of n: 193.4
## 95% Predicted Confidence Interval: [191.5, 195.4]G*Power gives estimate of 193 per group.
t-tests
Estimate sample size () per group in independent samples -test, one-tailed, medium effect size (), , 95% power (), equal sample sizes ().
##
## Execution time (H:M:S): 00:00:12
## Design conditions:
##
## # A tibble: 1 × 5
## n d two.tailed sig.level power
## <dbl> <dbl> <lgl> <dbl> <dbl>
## 1 NA 0.5 FALSE 0.05 0.95
##
## Estimate of n: 86.9
## 95% Predicted Confidence Interval: [85.2, 88.5]G*power estimate is 88 per group, Spower estimate is
86.9348 with the 95% CI [85.2011, 88.4739].
Example 19.3; Paired samples t-test
Paired-samples -test, assuming the generated difference is the repeated measures Cohen’s (e.g., were the unadjusted , while , then this results in the repeated ).
##
## Execution time (H:M:S): 00:00:22
## Design conditions:
##
## # A tibble: 1 × 4
## d type sig.level power
## <dbl> <chr> <dbl> <lgl>
## 1 0.42164 paired 0.05 NA
##
## Estimate of power: 0.840
## 95% Confidence Interval: [0.837, 0.843]G*power gives power estimate of .832, though Cohen reported a value closer to .840. When this leads to
##
## Execution time (H:M:S): 00:00:22
## Design conditions:
##
## # A tibble: 1 × 4
## d type sig.level power
## <dbl> <chr> <dbl> <lgl>
## 1 0.28284 paired 0.05 NA
##
## Estimate of power: 0.508
## 95% Confidence Interval: [0.503, 0.512]In this case G*Power 3.1 gives the estimate .500. To answer the
question “How many subjects would we need to arrive at a power of about
0.832114 in a two-group design?” this is specified within
Spower() and where n is set to NA
and Spower() is passed an interval argument,
or interval() is passed directly to the n
element in the experiment.
##
## Execution time (H:M:S): 00:00:21
## Design conditions:
##
## # A tibble: 1 × 5
## n d type sig.level power
## <dbl> <dbl> <chr> <dbl> <dbl>
## 1 NA 0.28284 paired 0.05 0.83211
##
## Estimate of n: 215.9
## 95% Predicted Confidence Interval: [213.7, 218.1]G*power reports that around pairs are required, though this is estimated visually using interpolation.
Example 20.3; One-sample t-test
Evaluating the hypotheses for the mean expression
using a one-sample -test. The following estimates given a one-tailed to achieve .
##
## Execution time (H:M:S): 00:00:09
## Design conditions:
##
## # A tibble: 1 × 6
## n d type two.tailed sig.level power
## <dbl> <dbl> <chr> <lgl> <dbl> <dbl>
## 1 NA 0.625 one.sample FALSE 0.05 0.95
##
## Estimate of n: 28.7
## 95% Predicted Confidence Interval: [28.0, 29.4]G*power gives sample size of . Similarly, though with different inputs.
##
## Execution time (H:M:S): 00:00:14
## Design conditions:
##
## # A tibble: 1 × 5
## n d type sig.level power
## <dbl> <dbl> <chr> <dbl> <dbl>
## 1 NA 0.1 one.sample 0.01 0.9
##
## Estimate of n: 1509.8
## 95% Predicted Confidence Interval: [1489.5, 1529.2]G*power gives sample size of .
Wilcoxon tests
Example 22.3; One-sample test with normal distribution
Same as Example 22.1 above.
p_wilcox.test(n=649, d=.1, type='one.sample', two.tailed=FALSE) |> Spower()##
## Execution time (H:M:S): 00:00:21
## Design conditions:
##
## # A tibble: 1 × 6
## n d type two.tailed sig.level power
## <dbl> <dbl> <chr> <lgl> <dbl> <lgl>
## 1 649 0.1 one.sample FALSE 0.05 NA
##
## Estimate of power: 0.799
## 95% Confidence Interval: [0.791, 0.806]G*power 3.1 provides a power estimate of .800, agreeing with
Spower.
Similarly, assuming that the distribution for the one-sample followed
a Laplace distribution, and that
were used instead. This requires defining an alternative parent
distribution, which below uses the rlaplace function from
the extraDistr package.
library(extraDistr)
parent1 <- function(n, d) extraDistr::rlaplace(n, mu=d, sigma=sqrt(1/2))
# properties of sampled distribution
descript(parent1(n=100000, d=0.8))## # A tibble: 1 × 14
## VARS n miss mean trimmed sd mad skewness kurtosis min Q_25
## <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 df 100000 NA 0.800 0.799 0.999 0.727 0.00702 3.06 -7.94 0.309
## # ℹ 3 more variables: Q_50 <dbl>, Q_75 <dbl>, max <dbl>
p_wilcox.test(n=11, d=.8, type='one.sample', two.tailed=FALSE, parent1 = parent1) |> Spower()##
## Execution time (H:M:S): 00:00:04
## Design conditions:
##
## # A tibble: 1 × 6
## n d type two.tailed sig.level power
## <dbl> <dbl> <chr> <lgl> <dbl> <lgl>
## 1 11 0.8 one.sample FALSE 0.05 NA
##
## Estimate of power: 0.815
## 95% Confidence Interval: [0.807, 0.822]Interestingly, G*power 3.1 reports this power to be 0.830.
Two-sample test with Laplace distributions
Two-sample Wilcoxon test comparing Laplace distributions with different central tendencies.
library(extraDistr)
parent1 <- function(n, d) extraDistr::rlaplace(n, mu=d, sigma=sqrt(1/2))
parent2 <- function(n, d) extraDistr::rlaplace(n, sigma=sqrt(1/2))
# properties of sampled distributions
descript(parent1(n=100000, d=0.375))## # A tibble: 1 × 14
## VARS n miss mean trimmed sd mad skewness kurtosis min Q_25
## <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 df 100000 NA 0.381 0.382 0.997 0.725 0.0256 3.02 -6.07 -0.107
## # ℹ 3 more variables: Q_50 <dbl>, Q_75 <dbl>, max <dbl>
descript(parent1(n=100000, d=0))## # A tibble: 1 × 14
## VARS n miss mean trimmed sd mad skewness kurtosis min Q_25
## <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 df 100000 NA 0.00267 0.00272 1.000 0.726 0.00490 2.86 -8.77 -0.485
## # ℹ 3 more variables: Q_50 <dbl>, Q_75 <dbl>, max <dbl>
nr <- 134/67
p_wilcox.test(n=67, n2_n1=nr, d=0.375, parent1=parent1, parent2=parent2) |>
Spower()##
## Execution time (H:M:S): 00:00:10
## Design conditions:
##
## # A tibble: 1 × 4
## n d sig.level power
## <dbl> <dbl> <dbl> <lgl>
## 1 67 0.375 0.05 NA
##
## Estimate of power: 0.851
## 95% Confidence Interval: [0.844, 0.858]Unlike before with the Laplace distribution, G*power 3.1 seems to
agree with Spower, where a power of .847 is reported. This
seems to raise questions about the consistency of the results.
Example 23.3: Paired-samples test with Laplace distributions
Finally, paired-samples approach using Wilcoxon test with .
parent1 <- function(n, d) extraDistr::rlaplace(n, mu=d, sigma=sqrt(1/2))
parent2 <- function(n, d) extraDistr::rlaplace(n, sigma=sqrt(1/2))
descript(parent1(n=100000, d=1.13842))## # A tibble: 1 × 14
## VARS n miss mean trimmed sd mad skewness kurtosis min Q_25
## <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 df 100000 NA 1.14 1.14 0.996 0.727 0.0158 2.93 -7.46 0.653
## # ℹ 3 more variables: Q_50 <dbl>, Q_75 <dbl>, max <dbl>
descript(parent1(n=100000, d=0))## # A tibble: 1 × 14
## VARS n miss mean trimmed sd mad skewness kurtosis min Q_25
## <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 df 1e5 NA -0.00205 -0.00284 1.00 0.722 0.0147 3.16 -7.60 -0.490
## # ℹ 3 more variables: Q_50 <dbl>, Q_75 <dbl>, max <dbl>
p_wilcox.test(n=10*2, d=1.13842, type = 'paired',
parent1=parent1, parent2=parent2) |> Spower()##
## Execution time (H:M:S): 00:00:04
## Design conditions:
##
## # A tibble: 1 × 4
## d type sig.level power
## <dbl> <chr> <dbl> <lgl>
## 1 1.1384 paired 0.05 NA
##
## Estimate of power: 0.933
## 95% Confidence Interval: [0.928, 0.938]Again, the simulation approach and G*power 3.1 differ in their outputs, where in G*power 3.1 the reported power is 0.853.