bfactor fits a confirmatory maximum likelihood two-tier/bifactor/testlet model to
dichotomous and polytomous data under the item response theory paradigm.
The IRT models are fit using a dimensional reduction EM algorithm so that regardless
of the number of specific factors estimated the model only uses the number of
factors in the second-tier structure plus 1. For the bifactor model the maximum
number of dimensions is only 2 since the second-tier only consists of a
ubiquitous unidimensional factor. See mirt for appropriate methods to be used
on the objects returned from the estimation.
Arguments
- data
a
matrixordata.framethat consists of numerically ordered data, organized in the form of integers, with missing data coded asNA- model
a numeric vector specifying which factor loads on which item. For example, if for a 4 item test with two specific factors, the first specific factor loads on the first two items and the second specific factor on the last two, then the vector is
c(1,1,2,2). For items that should only load on the second-tier factors (have no specific component)NAvalues may be used as place-holders. These numbers will be translated into a format suitable formirt.model(), combined with the definition inmodel2, with the letter 'S' added to the respective factor numberAlternatively, input can be specified using the
mirt.modelsyntax with the restriction that each item must load on exactly one specific factor (or no specific factors, if it is only predicted by the general factor specified inmodel2)- model2
a two-tier model specification object defined by
mirt.model()or a string to be passed tomirt.model. By default the model will fit a unidimensional model in the second-tier, and therefore be equivalent to the bifactor model- group
a factor variable indicating group membership used for multiple group analyses
- quadpts
number of quadrature nodes to use after accounting for the reduced number of dimensions. Scheme is the same as the one used in
mirt, however it is in regards to the reduced dimensions (e.g., a bifactor model has 2 dimensions to be integrated)- invariance
see
multipleGroupfor details, however, the specific factor variances and means will be constrained according to the dimensional reduction algorithm- ...
additional arguments to be passed to the estimation engine. See
mirtfor more details and examples
Value
function returns an object of class SingleGroupClass
(SingleGroupClass-class) or MultipleGroupClass(MultipleGroupClass-class).
Details
bfactor follows the item factor analysis strategy explicated by
Gibbons and Hedeker (1992), Gibbons et al. (2007), and Cai (2010).
Nested models may be compared via an approximate
chi-squared difference test or by a reduction in AIC or BIC (accessible via
anova). See mirt for more details regarding the
IRT estimation approach used in this package.
The two-tier model has a specific block diagonal covariance structure between the primary and secondary latent traits. Namely, the secondary latent traits are assumed to be orthogonal to all traits and have a fixed variance of 1, while the primary traits can be organized to vary and covary with other primary traits in the model.
$$\Sigma_{two-tier} = \left(\begin{array}{cc} G & 0 \\ 0 & diag(S) \end{array} \right)$$
The bifactor model is a special case of the two-tier model when \(G\) above is a 1x1 matrix, and therefore only 1 primary factor is being modeled. Evaluation of the numerical integrals for the two-tier model requires only \(ncol(G) + 1\) dimensions for integration since the \(S\) second order (or 'specific') factors require only 1 integration grid due to the dimension reduction technique.
Note: for multiple group two-tier analyses only the second-tier means and variances should be freed since the specific factors are not treated independently due to the dimension reduction technique.
References
Cai, L. (2010). A two-tier full-information item factor analysis model with applications. Psychometrika, 75, 581-612.
Chalmers, R., P. (2012). mirt: A Multidimensional Item Response Theory Package for the R Environment. Journal of Statistical Software, 48(6), 1-29. doi:10.18637/jss.v048.i06
Bradlow, E.T., Wainer, H., & Wang, X. (1999). A Bayesian random effects model for testlets. Psychometrika, 64, 153-168.
Gibbons, R. D., & Hedeker, D. R. (1992). Full-information Item Bi-Factor Analysis. Psychometrika, 57, 423-436.
Gibbons, R. D., Darrell, R. B., Hedeker, D., Weiss, D. J., Segawa, E., Bhaumik, D. K., Kupfer, D. J., Frank, E., Grochocinski, V. J., & Stover, A. (2007). Full-Information item bifactor analysis of graded response data. Applied Psychological Measurement, 31, 4-19.
Wainer, H., Bradlow, E.T., & Wang, X. (2007). Testlet response theory and its applications. New York, NY: Cambridge University Press.
Author
Phil Chalmers rphilip.chalmers@gmail.com
Examples
# \donttest{
### load SAT12 and compute bifactor model with 3 specific factors
data(SAT12)
data <- key2binary(SAT12,
key = c(1,4,5,2,3,1,2,1,3,1,2,4,2,1,5,3,4,4,1,4,3,3,4,1,3,5,1,3,1,5,4,5))
specific <- c(2,3,2,3,3,2,1,2,1,1,1,3,1,3,1,2,1,1,3,3,1,1,3,1,3,3,1,3,2,3,1,2)
mod1 <- bfactor(data, specific)
summary(mod1)
#> G S1 S2 S3 h2
#> Item.1 0.4078 0.0000 0.2273 0.0000 0.21799
#> Item.2 0.6195 0.0000 0.0000 0.3391 0.49881
#> Item.3 0.5573 0.0000 -0.0744 0.0000 0.31612
#> Item.4 0.2808 0.0000 0.0000 0.3101 0.17503
#> Item.5 0.4779 0.0000 0.0000 0.2544 0.29311
#> Item.6 0.5341 0.0000 0.2724 0.0000 0.35948
#> Item.7 0.4741 0.4210 0.0000 0.0000 0.40199
#> Item.8 0.3537 0.0000 0.2732 0.0000 0.19971
#> Item.9 0.2181 0.5321 0.0000 0.0000 0.33068
#> Item.10 0.4849 0.3792 0.0000 0.0000 0.37895
#> Item.11 0.6442 0.3321 0.0000 0.0000 0.52536
#> Item.12 0.0699 0.0000 0.0000 0.1592 0.03023
#> Item.13 0.5219 0.2743 0.0000 0.0000 0.34764
#> Item.14 0.4791 0.0000 0.0000 0.4563 0.43776
#> Item.15 0.5985 0.2402 0.0000 0.0000 0.41590
#> Item.16 0.3885 0.0000 0.2049 0.0000 0.19292
#> Item.17 0.6636 0.1177 0.0000 0.0000 0.45423
#> Item.18 0.7163 0.0775 0.0000 0.0000 0.51903
#> Item.19 0.4520 0.0000 0.0000 0.0198 0.20466
#> Item.20 0.6578 0.0000 0.0000 0.1792 0.46478
#> Item.21 0.2806 0.3451 0.0000 0.0000 0.19787
#> Item.22 0.7025 -0.0281 0.0000 0.0000 0.49425
#> Item.23 0.3236 0.0000 0.0000 0.2657 0.17536
#> Item.24 0.5848 0.1030 0.0000 0.0000 0.35266
#> Item.25 0.3732 0.0000 0.0000 0.3297 0.24799
#> Item.26 0.6430 0.0000 0.0000 0.2124 0.45854
#> Item.27 0.7374 0.1554 0.0000 0.0000 0.56792
#> Item.28 0.5256 0.0000 0.0000 0.0758 0.28199
#> Item.29 0.4185 0.0000 0.7071 0.0000 0.67516
#> Item.30 0.2455 0.0000 0.0000 -0.0959 0.06946
#> Item.31 0.8333 -0.0872 0.0000 0.0000 0.70202
#> Item.32 0.0780 0.0000 0.0165 0.0000 0.00635
#>
#> SS loadings: 8.435 1.03 0.748 0.781
#> Proportion Var: 0.264 0.032 0.023 0.024
#>
#> Factor correlations:
#>
#> G S1 S2 S3
#> G 1
#> S1 0 1
#> S2 0 0 1
#> S3 0 0 0 1
itemplot(mod1, 18, drop.zeros = TRUE) #drop the zero slopes to allow plotting
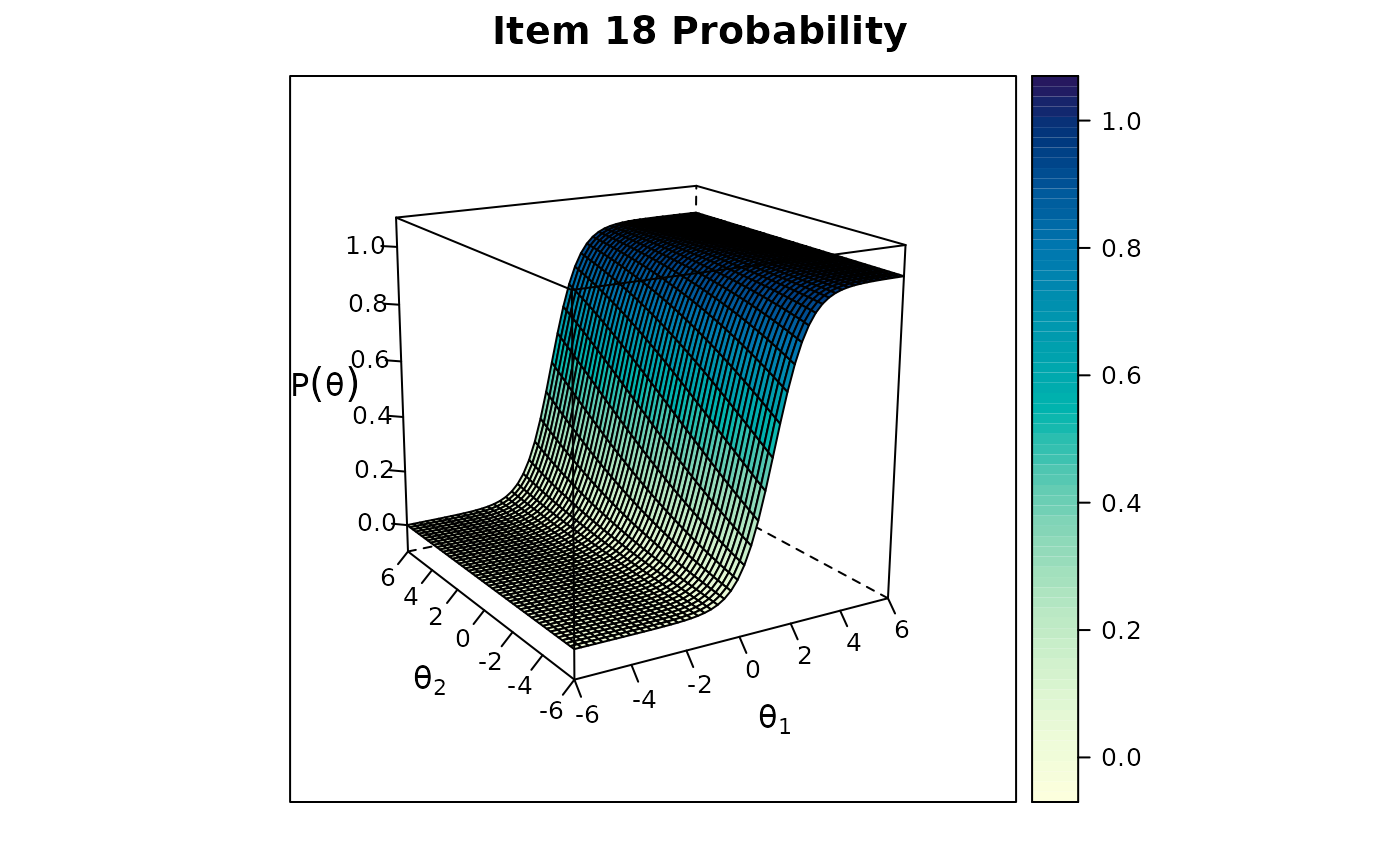 # alternative model definition via ?mirt.model syntax
specific2 <- "S1 = 7,9,10,11,13,15,17,18,21,22,24,27,31
S2 = 1,3,6,8,16,29,32
S3 = 2,4,5,12,14,19,20,23,25,26,28,30"
mod2 <- bfactor(data, specific2)
anova(mod1, mod2) # same
#> AIC SABIC HQ BIC logLik X2 df p
#> mod1 19062.1 19179.44 19226.42 19484.21 -9435.052
#> mod2 19062.1 19179.44 19226.42 19484.21 -9435.052 0 0 NaN
# also equivalent using item names instead (not run)
specific3 <- "S1 = Item.7, Item.9, Item.10, Item.11, Item.13, Item.15,
Item.17, Item.18, Item.21, Item.22, Item.24, Item.27, Item.31
S2 = Item.1, Item.3, Item.6, Item.8, Item.16, Item.29, Item.32
S3 = Item.2, Item.4, Item.5, Item.12, Item.14, Item.19,
Item.20, Item.23, Item.25, Item.26, Item.28, Item.30"
# mod3 <- bfactor(data, specific3)
# anova(mod1, mod2, mod3) # all same
### Try with fixed guessing parameters added
guess <- rep(.1,32)
mod2 <- bfactor(data, specific, guess = guess)
coef(mod2)
#> $Item.1
#> a1 a2 a3 a4 d g u
#> par 1.225 0 0.624 0 -1.822 0.1 1
#>
#> $Item.2
#> a1 a2 a3 a4 d g u
#> par 1.721 0 0 0.954 0.171 0.1 1
#>
#> $Item.3
#> a1 a2 a3 a4 d g u
#> par 2.415 0 -0.459 0 -2.602 0.1 1
#>
#> $Item.4
#> a1 a2 a3 a4 d g u
#> par 0.745 0 0 0.695 -0.989 0.1 1
#>
#> $Item.5
#> a1 a2 a3 a4 d g u
#> par 1.048 0 0 0.603 0.419 0.1 1
#>
#> $Item.6
#> a1 a2 a3 a4 d g u
#> par 3.06 0 0.501 0 -5.002 0.1 1
#>
#> $Item.7
#> a1 a2 a3 a4 d g u
#> par 1.121 0.839 0 0 1.373 0.1 1
#>
#> $Item.8
#> a1 a2 a3 a4 d g u
#> par 1.956 0 1.443 0 -3.772 0.1 1
#>
#> $Item.9
#> a1 a2 a3 a4 d g u
#> par 0.512 1.236 0 0 2.484 0.1 1
#>
#> $Item.10
#> a1 a2 a3 a4 d g u
#> par 1.68 1.506 0 0 -1.031 0.1 1
#>
#> $Item.11
#> a1 a2 a3 a4 d g u
#> par 1.655 0.842 0 0 5.441 0.1 1
#>
#> $Item.12
#> a1 a2 a3 a4 d g u
#> par 0.129 0 0 0.364 -0.641 0.1 1
#>
#> $Item.13
#> a1 a2 a3 a4 d g u
#> par 1.183 0.477 0 0 0.679 0.1 1
#>
#> $Item.14
#> a1 a2 a3 a4 d g u
#> par 1.125 0 0 1.058 1.164 0.1 1
#>
#> $Item.15
#> a1 a2 a3 a4 d g u
#> par 1.435 0.317 0 0 1.863 0.1 1
#>
#> $Item.16
#> a1 a2 a3 a4 d g u
#> par 0.95 0 0.573 0 -0.783 0.1 1
#>
#> $Item.17
#> a1 a2 a3 a4 d g u
#> par 1.547 0.059 0 0 4.112 0.1 1
#>
#> $Item.18
#> a1 a2 a3 a4 d g u
#> par 2.731 0.094 0 0 -1.808 0.1 1
#>
#> $Item.19
#> a1 a2 a3 a4 d g u
#> par 0.918 0 0 0.101 -0.001 0.1 1
#>
#> $Item.20
#> a1 a2 a3 a4 d g u
#> par 1.456 0 0 0.593 2.501 0.1 1
#>
#> $Item.21
#> a1 a2 a3 a4 d g u
#> par 0.596 0.493 0 0 2.49 0.1 1
#>
#> $Item.22
#> a1 a2 a3 a4 d g u
#> par 1.554 -0.242 0 0 3.428 0.1 1
#>
#> $Item.23
#> a1 a2 a3 a4 d g u
#> par 0.908 0 0 0.766 -1.488 0.1 1
#>
#> $Item.24
#> a1 a2 a3 a4 d g u
#> par 1.379 0.001 0 0 1.132 0.1 1
#>
#> $Item.25
#> a1 a2 a3 a4 d g u
#> par 1.03 0 0 1.094 -1.164 0.1 1
#>
#> $Item.26
#> a1 a2 a3 a4 d g u
#> par 1.985 0 0 0.747 -0.663 0.1 1
#>
#> $Item.27
#> a1 a2 a3 a4 d g u
#> par 1.909 0.348 0 0 2.642 0.1 1
#>
#> $Item.28
#> a1 a2 a3 a4 d g u
#> par 1.213 0 0 0.142 -0.097 0.1 1
#>
#> $Item.29
#> a1 a2 a3 a4 d g u
#> par 1.938 0 2.339 0 -2.209 0.1 1
#>
#> $Item.30
#> a1 a2 a3 a4 d g u
#> par 0.479 0 0 -0.128 -0.527 0.1 1
#>
#> $Item.31
#> a1 a2 a3 a4 d g u
#> par 3.173 -0.82 0 0 3.316 0.1 1
#>
#> $Item.32
#> a1 a2 a3 a4 d g u
#> par 0.534 0 -0.053 0 -2.786 0.1 1
#>
#> $GroupPars
#> MEAN_1 MEAN_2 MEAN_3 MEAN_4 COV_11 COV_21 COV_31 COV_41 COV_22 COV_32
#> par 0 0 0 0 1 0 0 0 1 0
#> COV_42 COV_33 COV_43 COV_44
#> par 0 1 0 1
#>
anova(mod1, mod2)
#> AIC SABIC HQ BIC logLik X2 df p
#> mod1 19062.10 19179.44 19226.42 19484.21 -9435.052
#> mod2 19009.55 19126.88 19173.87 19431.65 -9408.775 52.553 0 NaN
## don't estimate specific factor for item 32
specific[32] <- NA
mod3 <- bfactor(data, specific)
anova(mod3, mod1)
#> AIC SABIC HQ BIC logLik X2 df p
#> mod3 19060.12 19176.23 19222.73 19477.83 -9435.062
#> mod1 19062.10 19179.44 19226.42 19484.21 -9435.052 0.02 1 0.886
# same, but with syntax (not run)
specific3 <- "S1 = 7,9,10,11,13,15,17,18,21,22,24,27,31
S2 = 1,3,6,8,16,29
S3 = 2,4,5,12,14,19,20,23,25,26,28,30"
# mod3b <- bfactor(data, specific3)
# anova(mod3b)
#########
# mixed itemtype example
# simulate data
a <- matrix(c(
1,0.5,NA,
1,0.5,NA,
1,0.5,NA,
1,0.5,NA,
1,0.5,NA,
1,0.5,NA,
1,0.5,NA,
1,NA,0.5,
1,NA,0.5,
1,NA,0.5,
1,NA,0.5,
1,NA,0.5,
1,NA,0.5,
1,NA,0.5),ncol=3,byrow=TRUE)
d <- matrix(c(
-1.0,NA,NA,
-1.5,NA,NA,
1.5,NA,NA,
0.0,NA,NA,
2.5,1.0,-1,
3.0,2.0,-0.5,
3.0,2.0,-0.5,
3.0,2.0,-0.5,
2.5,1.0,-1,
2.0,0.0,NA,
-1.0,NA,NA,
-1.5,NA,NA,
1.5,NA,NA,
0.0,NA,NA),ncol=3,byrow=TRUE)
items <- rep('2PL', 14)
items[5:10] <- 'graded'
sigma <- diag(3)
dataset <- simdata(a,d,5000,itemtype=items,sigma=sigma)
itemstats(dataset)
#> $overall
#> N mean_total.score sd_total.score ave.r sd.r alpha SEM.alpha
#> 5000 15.116 4.495 0.175 0.034 0.733 2.323
#>
#> $itemstats
#> N mean sd total.r total.r_if_rm alpha_if_rm
#> Item_1 5000 0.311 0.463 0.434 0.345 0.720
#> Item_2 5000 0.229 0.420 0.386 0.302 0.724
#> Item_3 5000 0.770 0.421 0.378 0.293 0.724
#> Item_4 5000 0.493 0.500 0.455 0.360 0.718
#> Item_5 5000 1.893 0.975 0.585 0.414 0.711
#> Item_6 5000 2.150 0.883 0.537 0.374 0.716
#> Item_7 5000 2.168 0.881 0.538 0.376 0.715
#> Item_8 5000 2.138 0.889 0.549 0.388 0.714
#> Item_9 5000 1.857 0.978 0.586 0.413 0.711
#> Item_10 5000 1.327 0.746 0.523 0.387 0.713
#> Item_11 5000 0.304 0.460 0.427 0.338 0.721
#> Item_12 5000 0.234 0.424 0.403 0.320 0.723
#> Item_13 5000 0.752 0.432 0.417 0.333 0.722
#> Item_14 5000 0.491 0.500 0.457 0.362 0.718
#>
#> $proportions
#> 0 1 2 3
#> Item_1 0.689 0.311 NA NA
#> Item_2 0.771 0.229 NA NA
#> Item_3 0.230 0.770 NA NA
#> Item_4 0.507 0.493 NA NA
#> Item_5 0.114 0.194 0.378 0.314
#> Item_6 0.079 0.089 0.434 0.397
#> Item_7 0.079 0.083 0.431 0.408
#> Item_8 0.081 0.093 0.433 0.393
#> Item_9 0.115 0.214 0.370 0.301
#> Item_10 0.168 0.337 0.495 NA
#> Item_11 0.696 0.304 NA NA
#> Item_12 0.766 0.234 NA NA
#> Item_13 0.248 0.752 NA NA
#> Item_14 0.509 0.491 NA NA
#>
specific <- "S1 = 1-7
S2 = 8-14"
simmod <- bfactor(dataset, specific)
coef(simmod, simplify=TRUE)
#> $items
#> a1 a2 a3 d g u d1 d2 d3
#> Item_1 1.032 0.505 0.000 -1.003 0 1 NA NA NA
#> Item_2 0.953 0.474 0.000 -1.477 0 1 NA NA NA
#> Item_3 0.868 0.494 0.000 1.442 0 1 NA NA NA
#> Item_4 1.002 0.406 0.000 -0.037 0 1 NA NA NA
#> Item_5 1.040 0.578 0.000 NA NA NA 2.536 1.029 -0.998
#> Item_6 0.963 0.465 0.000 NA NA NA 2.899 1.928 -0.514
#> Item_7 0.969 0.504 0.000 NA NA NA 2.929 2.009 -0.462
#> Item_8 1.011 0.000 0.680 NA NA NA 2.993 1.974 -0.563
#> Item_9 1.023 0.000 0.469 NA NA NA 2.471 0.888 -1.054
#> Item_10 1.002 0.000 0.461 NA NA NA 1.951 -0.023 NA
#> Item_11 1.001 0.000 0.543 -1.041 0 1 NA NA NA
#> Item_12 1.039 0.000 0.603 -1.502 0 1 NA NA NA
#> Item_13 1.033 0.000 0.429 1.377 0 1 NA NA NA
#> Item_14 1.017 0.000 0.401 -0.046 0 1 NA NA NA
#>
#> $means
#> G S1 S2
#> 0 0 0
#>
#> $cov
#> G S1 S2
#> G 1 0 0
#> S1 0 1 0
#> S2 0 0 1
#>
#########
# General testlet response model (Wainer, 2007)
# simulate data
set.seed(1234)
a <- matrix(0, 12, 4)
a[,1] <- rlnorm(12, .2, .3)
ind <- 1
for(i in 1:3){
a[ind:(ind+3),i+1] <- a[ind:(ind+3),1]
ind <- ind+4
}
print(a)
#> [,1] [,2] [,3] [,4]
#> [1,] 0.8503394 0.8503394 0.000000 0.0000000
#> [2,] 1.3274088 1.3274088 0.000000 0.0000000
#> [3,] 1.6910208 1.6910208 0.000000 0.0000000
#> [4,] 0.6042850 0.6042850 0.000000 0.0000000
#> [5,] 1.3892130 0.0000000 1.389213 0.0000000
#> [6,] 1.4216480 0.0000000 1.421648 0.0000000
#> [7,] 1.0279618 0.0000000 1.027962 0.0000000
#> [8,] 1.0366667 0.0000000 1.036667 0.0000000
#> [9,] 1.0311394 0.0000000 0.000000 1.0311394
#> [10,] 0.9351846 0.0000000 0.000000 0.9351846
#> [11,] 1.0584888 0.0000000 0.000000 1.0584888
#> [12,] 0.9052755 0.0000000 0.000000 0.9052755
d <- rnorm(12, 0, .5)
sigma <- diag(c(1, .5, 1, .5))
dataset <- simdata(a,d,2000,itemtype=rep('2PL', 12),sigma=sigma)
itemstats(dataset)
#> $overall
#> N mean_total.score sd_total.score ave.r sd.r alpha SEM.alpha
#> 2000 6 2.929 0.175 0.068 0.717 1.558
#>
#> $itemstats
#> N mean sd total.r total.r_if_rm alpha_if_rm
#> Item_1 2000 0.426 0.495 0.438 0.287 0.708
#> Item_2 2000 0.502 0.500 0.560 0.425 0.689
#> Item_3 2000 0.571 0.495 0.575 0.445 0.686
#> Item_4 2000 0.502 0.500 0.383 0.224 0.716
#> Item_5 2000 0.464 0.499 0.549 0.413 0.690
#> Item_6 2000 0.436 0.496 0.561 0.428 0.688
#> Item_7 2000 0.440 0.497 0.500 0.356 0.698
#> Item_8 2000 0.693 0.462 0.474 0.339 0.701
#> Item_9 2000 0.511 0.500 0.481 0.334 0.701
#> Item_10 2000 0.456 0.498 0.465 0.316 0.704
#> Item_11 2000 0.458 0.498 0.459 0.309 0.705
#> Item_12 2000 0.540 0.498 0.475 0.327 0.702
#>
#> $proportions
#> 0 1
#> Item_1 0.575 0.426
#> Item_2 0.498 0.502
#> Item_3 0.430 0.571
#> Item_4 0.498 0.502
#> Item_5 0.536 0.464
#> Item_6 0.564 0.436
#> Item_7 0.559 0.440
#> Item_8 0.308 0.693
#> Item_9 0.488 0.511
#> Item_10 0.543 0.456
#> Item_11 0.541 0.458
#> Item_12 0.460 0.540
#>
# estimate by applying constraints and freeing the latent variances
specific <- "S1 = 1-4
S2 = 5-8
S3 = 9-12"
model <- "G = 1-12
CONSTRAIN = (1, a1, a2), (2, a1, a2), (3, a1, a2), (4, a1, a2),
(5, a1, a3), (6, a1, a3), (7, a1, a3), (8, a1, a3),
(9, a1, a4), (10, a1, a4), (11, a1, a4), (12, a1, a4)
COV = S1*S1, S2*S2, S3*S3"
simmod <- bfactor(dataset, specific, model)
coef(simmod, simplify=TRUE)
#> $items
#> a1 a2 a3 a4 d g u
#> Item_1 0.794 0.794 0.000 0.000 -0.359 0 1
#> Item_2 1.544 1.544 0.000 0.000 0.011 0 1
#> Item_3 1.762 1.762 0.000 0.000 0.479 0 1
#> Item_4 0.544 0.544 0.000 0.000 0.011 0 1
#> Item_5 1.386 0.000 1.386 0.000 -0.244 0 1
#> Item_6 1.497 0.000 1.497 0.000 -0.449 0 1
#> Item_7 0.853 0.000 0.853 0.000 -0.312 0 1
#> Item_8 0.953 0.000 0.953 0.000 1.101 0 1
#> Item_9 0.981 0.000 0.000 0.981 0.058 0 1
#> Item_10 0.913 0.000 0.000 0.913 -0.217 0 1
#> Item_11 0.868 0.000 0.000 0.868 -0.204 0 1
#> Item_12 0.966 0.000 0.000 0.966 0.206 0 1
#>
#> $means
#> G S1 S2 S3
#> 0 0 0 0
#>
#> $cov
#> G S1 S2 S3
#> G 1 0.000 0.000 0.000
#> S1 0 0.452 0.000 0.000
#> S2 0 0.000 1.135 0.000
#> S3 0 0.000 0.000 0.432
#>
# Constrained testlet model (Bradlow, 1999)
model2 <- "G = 1-12
CONSTRAIN = (1, a1, a2), (2, a1, a2), (3, a1, a2), (4, a1, a2),
(5, a1, a3), (6, a1, a3), (7, a1, a3), (8, a1, a3),
(9, a1, a4), (10, a1, a4), (11, a1, a4), (12, a1, a4),
(GROUP, COV_22, COV_33, COV_44)
COV = S1*S1, S2*S2, S3*S3"
simmod2 <- bfactor(dataset, specific, model2)
coef(simmod2, simplify=TRUE)
#> $items
#> a1 a2 a3 a4 d g u
#> Item_1 0.744 0.744 0.000 0.000 -0.360 0 1
#> Item_2 1.453 1.453 0.000 0.000 0.010 0 1
#> Item_3 1.664 1.664 0.000 0.000 0.482 0 1
#> Item_4 0.509 0.509 0.000 0.000 0.011 0 1
#> Item_5 1.541 0.000 1.541 0.000 -0.241 0 1
#> Item_6 1.670 0.000 1.670 0.000 -0.445 0 1
#> Item_7 0.968 0.000 0.968 0.000 -0.313 0 1
#> Item_8 1.075 0.000 1.075 0.000 1.098 0 1
#> Item_9 0.927 0.000 0.000 0.927 0.059 0 1
#> Item_10 0.854 0.000 0.000 0.854 -0.218 0 1
#> Item_11 0.813 0.000 0.000 0.813 -0.205 0 1
#> Item_12 0.908 0.000 0.000 0.908 0.207 0 1
#>
#> $means
#> G S1 S2 S3
#> 0 0 0 0
#>
#> $cov
#> G S1 S2 S3
#> G 1 0.000 0.000 0.000
#> S1 0 0.667 0.000 0.000
#> S2 0 0.000 0.667 0.000
#> S3 0 0.000 0.000 0.667
#>
anova(simmod2, simmod)
#> AIC SABIC HQ BIC logLik X2 df p
#> simmod2 30256.59 30317.19 30308.00 30396.61 -15103.3
#> simmod 30248.79 30314.24 30304.32 30400.02 -15097.4 11.795 2 0.003
#########
# Two-tier model
# simulate data
set.seed(1234)
a <- matrix(c(
0,1,0.5,NA,NA,
0,1,0.5,NA,NA,
0,1,0.5,NA,NA,
0,1,0.5,NA,NA,
0,1,0.5,NA,NA,
0,1,NA,0.5,NA,
0,1,NA,0.5,NA,
0,1,NA,0.5,NA,
1,0,NA,0.5,NA,
1,0,NA,0.5,NA,
1,0,NA,0.5,NA,
1,0,NA,NA,0.5,
1,0,NA,NA,0.5,
1,0,NA,NA,0.5,
1,0,NA,NA,0.5,
1,0,NA,NA,0.5),ncol=5,byrow=TRUE)
d <- matrix(rnorm(16))
items <- rep('2PL', 16)
sigma <- diag(5)
sigma[1,2] <- sigma[2,1] <- .4
dataset <- simdata(a,d,2000,itemtype=items,sigma=sigma)
itemstats(dataset)
#> $overall
#> N mean_total.score sd_total.score ave.r sd.r alpha SEM.alpha
#> 2000 7.086 3.077 0.108 0.058 0.662 1.79
#>
#> $itemstats
#> N mean sd total.r total.r_if_rm alpha_if_rm
#> Item_1 2000 0.288 0.453 0.378 0.241 0.650
#> Item_2 2000 0.571 0.495 0.422 0.276 0.646
#> Item_3 2000 0.705 0.456 0.381 0.245 0.650
#> Item_4 2000 0.133 0.340 0.289 0.183 0.656
#> Item_5 2000 0.601 0.490 0.393 0.246 0.650
#> Item_6 2000 0.587 0.492 0.419 0.274 0.646
#> Item_7 2000 0.379 0.485 0.444 0.304 0.642
#> Item_8 2000 0.378 0.485 0.400 0.256 0.649
#> Item_9 2000 0.386 0.487 0.392 0.246 0.650
#> Item_10 2000 0.322 0.467 0.400 0.261 0.648
#> Item_11 2000 0.402 0.490 0.455 0.315 0.640
#> Item_12 2000 0.318 0.466 0.414 0.278 0.646
#> Item_13 2000 0.368 0.482 0.423 0.281 0.645
#> Item_14 2000 0.498 0.500 0.424 0.277 0.646
#> Item_15 2000 0.669 0.471 0.394 0.254 0.649
#> Item_16 2000 0.482 0.500 0.444 0.300 0.642
#>
#> $proportions
#> 0 1
#> Item_1 0.713 0.288
#> Item_2 0.430 0.571
#> Item_3 0.295 0.705
#> Item_4 0.867 0.133
#> Item_5 0.400 0.601
#> Item_6 0.413 0.587
#> Item_7 0.621 0.379
#> Item_8 0.622 0.378
#> Item_9 0.614 0.386
#> Item_10 0.678 0.322
#> Item_11 0.598 0.402
#> Item_12 0.681 0.318
#> Item_13 0.632 0.368
#> Item_14 0.502 0.498
#> Item_15 0.330 0.669
#> Item_16 0.518 0.482
#>
specific <- "S1 = 1-5
S2 = 6-11
S3 = 12-16"
model <- '
G1 = 1-8
G2 = 9-16
COV = G1*G2'
# quadpts dropped for faster estimation, but not as precise
simmod <- bfactor(dataset, specific, model, quadpts = 9, TOL = 1e-3)
coef(simmod, simplify=TRUE)
#> $items
#> a1 a2 a3 a4 a5 d g u
#> Item_1 0.965 0.000 0.385 0.000 0.000 -1.100 0 1
#> Item_2 1.076 0.000 0.550 0.000 0.000 0.363 0 1
#> Item_3 0.898 0.000 0.592 0.000 0.000 1.068 0 1
#> Item_4 0.896 0.000 0.710 0.000 0.000 -2.293 0 1
#> Item_5 0.892 0.000 0.848 0.000 0.000 0.526 0 1
#> Item_6 1.013 0.000 0.000 0.413 0.000 0.435 0 1
#> Item_7 1.162 0.000 0.000 0.451 0.000 -0.639 0 1
#> Item_8 0.945 0.000 0.000 0.609 0.000 -0.623 0 1
#> Item_9 0.000 0.831 0.000 0.371 0.000 -0.544 0 1
#> Item_10 0.000 0.925 0.000 0.610 0.000 -0.926 0 1
#> Item_11 0.000 1.142 0.000 0.495 0.000 -0.517 0 1
#> Item_12 0.000 0.978 0.000 0.000 0.634 -0.964 0 1
#> Item_13 0.000 1.108 0.000 0.000 0.437 -0.694 0 1
#> Item_14 0.000 1.004 0.000 0.000 0.321 -0.012 0 1
#> Item_15 0.000 0.916 0.000 0.000 0.758 0.897 0 1
#> Item_16 0.000 1.020 0.000 0.000 0.650 -0.096 0 1
#>
#> $means
#> G1 G2 S1 S2 S3
#> 0 0 0 0 0
#>
#> $cov
#> G1 G2 S1 S2 S3
#> G1 1.000 0.412 0 0 0
#> G2 0.412 1.000 0 0 0
#> S1 0.000 0.000 1 0 0
#> S2 0.000 0.000 0 1 0
#> S3 0.000 0.000 0 0 1
#>
summary(simmod)
#> G1 G2 S1 S2 S3 h2
#> Item_1 0.484 0.000 0.193 0.000 0.000 0.271
#> Item_2 0.516 0.000 0.263 0.000 0.000 0.335
#> Item_3 0.446 0.000 0.294 0.000 0.000 0.285
#> Item_4 0.437 0.000 0.346 0.000 0.000 0.311
#> Item_5 0.425 0.000 0.404 0.000 0.000 0.343
#> Item_6 0.501 0.000 0.000 0.204 0.000 0.293
#> Item_7 0.551 0.000 0.000 0.214 0.000 0.349
#> Item_8 0.463 0.000 0.000 0.299 0.000 0.304
#> Item_9 0.000 0.431 0.000 0.192 0.000 0.222
#> Item_10 0.000 0.456 0.000 0.300 0.000 0.298
#> Item_11 0.000 0.541 0.000 0.235 0.000 0.348
#> Item_12 0.000 0.474 0.000 0.000 0.307 0.319
#> Item_13 0.000 0.533 0.000 0.000 0.210 0.329
#> Item_14 0.000 0.501 0.000 0.000 0.160 0.277
#> Item_15 0.000 0.441 0.000 0.000 0.365 0.328
#> Item_16 0.000 0.488 0.000 0.000 0.311 0.336
#>
#> SS loadings: 1.839 1.88 0.476 0.359 0.395
#> Proportion Var: 0.115 0.118 0.03 0.022 0.025
#>
#> Factor correlations:
#>
#> G1 G2 S1 S2 S3
#> G1 1.000
#> G2 0.412 1
#> S1 0.000 0 1
#> S2 0.000 0 0 1
#> S3 0.000 0 0 0 1
itemfit(simmod, QMC=TRUE)
#> item S_X2 df.S_X2 RMSEA.S_X2 p.S_X2
#> 1 Item_1 7.103 9 0.000 0.626
#> 2 Item_2 13.326 10 0.013 0.206
#> 3 Item_3 8.332 9 0.000 0.501
#> 4 Item_4 8.531 10 0.000 0.577
#> 5 Item_5 7.170 10 0.000 0.709
#> 6 Item_6 3.967 10 0.000 0.949
#> 7 Item_7 8.350 10 0.000 0.595
#> 8 Item_8 16.010 10 0.017 0.099
#> 9 Item_9 17.529 10 0.019 0.063
#> 10 Item_10 12.058 10 0.010 0.281
#> 11 Item_11 13.567 10 0.013 0.194
#> 12 Item_12 13.907 9 0.017 0.126
#> 13 Item_13 11.144 10 0.008 0.346
#> 14 Item_14 7.852 10 0.000 0.643
#> 15 Item_15 14.142 9 0.017 0.117
#> 16 Item_16 5.926 10 0.000 0.821
M2(simmod, QMC=TRUE)
#> M2 df p RMSEA RMSEA_5 RMSEA_95 SRMSR TLI CFI
#> stats 86.28163 87 0.5015988 0 0 0.01201603 0.01662365 1.000285 1
residuals(simmod, QMC=TRUE)
#> LD matrix (lower triangle) and standardized residual correlations (upper triangle)
#>
#> Upper triangle summary:
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> -0.046 -0.011 -0.002 -0.001 0.012 0.041
#>
#> Item_1 Item_2 Item_3 Item_4 Item_5 Item_6 Item_7 Item_8 Item_9 Item_10
#> Item_1 -0.011 -0.015 -0.007 0.016 0.003 0.006 -0.002 0.004 -0.009
#> Item_2 0.263 -0.002 0.005 -0.001 0.016 -0.022 0.021 -0.003 -0.006
#> Item_3 0.441 0.008 0.014 -0.001 0.029 -0.017 -0.011 -0.021 -0.028
#> Item_4 0.086 0.054 0.376 -0.014 -0.023 0.021 -0.004 0.020 -0.040
#> Item_5 0.514 0.004 0.003 0.386 -0.028 0.011 0.014 -0.021 0.013
#> Item_6 0.015 0.483 1.630 1.038 1.588 -0.022 -0.009 0.031 -0.004
#> Item_7 0.077 0.996 0.590 0.852 0.258 0.992 -0.007 -0.004 0.013
#> Item_8 0.012 0.858 0.264 0.039 0.377 0.154 0.094 -0.020 0.008
#> Item_9 0.033 0.017 0.863 0.803 0.890 1.974 0.037 0.808 0.001
#> Item_10 0.157 0.084 1.528 3.158 0.360 0.038 0.330 0.128 0.001
#> Item_11 0.510 0.125 2.195 0.231 0.004 0.215 1.049 0.004 0.031 0.754
#> Item_12 2.017 0.253 1.865 0.388 0.005 0.417 0.074 0.090 0.443 0.042
#> Item_13 0.470 2.122 0.125 0.271 0.881 0.264 0.310 4.304 0.009 0.059
#> Item_14 0.101 1.546 0.165 0.006 0.296 0.004 1.672 0.765 3.341 0.066
#> Item_15 0.822 0.257 0.011 0.442 0.443 0.113 0.526 0.297 2.306 0.044
#> Item_16 0.097 0.627 1.486 0.127 0.445 0.011 0.732 0.061 0.007 0.944
#> Item_11 Item_12 Item_13 Item_14 Item_15 Item_16
#> Item_1 -0.016 -0.032 0.015 -0.007 0.020 -0.007
#> Item_2 0.008 0.011 -0.033 -0.028 -0.011 0.018
#> Item_3 0.033 0.031 0.008 -0.009 -0.002 0.027
#> Item_4 -0.011 0.014 -0.012 -0.002 -0.015 -0.008
#> Item_5 -0.001 0.002 -0.021 -0.012 0.015 0.015
#> Item_6 0.010 0.014 -0.011 0.001 -0.008 0.002
#> Item_7 0.023 0.006 0.012 0.029 -0.016 0.019
#> Item_8 -0.001 0.007 -0.046 -0.020 0.012 0.006
#> Item_9 0.004 -0.015 -0.002 0.041 -0.034 -0.002
#> Item_10 -0.019 0.005 0.005 0.006 -0.005 0.022
#> Item_11 0.009 0.000 -0.025 0.039 -0.011
#> Item_12 0.149 -0.008 0.013 -0.002 -0.009
#> Item_13 0.000 0.140 0.013 -0.012 0.003
#> Item_14 1.269 0.338 0.336 -0.007 -0.023
#> Item_15 3.061 0.010 0.284 0.086 0.010
#> Item_16 0.250 0.153 0.015 1.026 0.190
# }
# alternative model definition via ?mirt.model syntax
specific2 <- "S1 = 7,9,10,11,13,15,17,18,21,22,24,27,31
S2 = 1,3,6,8,16,29,32
S3 = 2,4,5,12,14,19,20,23,25,26,28,30"
mod2 <- bfactor(data, specific2)
anova(mod1, mod2) # same
#> AIC SABIC HQ BIC logLik X2 df p
#> mod1 19062.1 19179.44 19226.42 19484.21 -9435.052
#> mod2 19062.1 19179.44 19226.42 19484.21 -9435.052 0 0 NaN
# also equivalent using item names instead (not run)
specific3 <- "S1 = Item.7, Item.9, Item.10, Item.11, Item.13, Item.15,
Item.17, Item.18, Item.21, Item.22, Item.24, Item.27, Item.31
S2 = Item.1, Item.3, Item.6, Item.8, Item.16, Item.29, Item.32
S3 = Item.2, Item.4, Item.5, Item.12, Item.14, Item.19,
Item.20, Item.23, Item.25, Item.26, Item.28, Item.30"
# mod3 <- bfactor(data, specific3)
# anova(mod1, mod2, mod3) # all same
### Try with fixed guessing parameters added
guess <- rep(.1,32)
mod2 <- bfactor(data, specific, guess = guess)
coef(mod2)
#> $Item.1
#> a1 a2 a3 a4 d g u
#> par 1.225 0 0.624 0 -1.822 0.1 1
#>
#> $Item.2
#> a1 a2 a3 a4 d g u
#> par 1.721 0 0 0.954 0.171 0.1 1
#>
#> $Item.3
#> a1 a2 a3 a4 d g u
#> par 2.415 0 -0.459 0 -2.602 0.1 1
#>
#> $Item.4
#> a1 a2 a3 a4 d g u
#> par 0.745 0 0 0.695 -0.989 0.1 1
#>
#> $Item.5
#> a1 a2 a3 a4 d g u
#> par 1.048 0 0 0.603 0.419 0.1 1
#>
#> $Item.6
#> a1 a2 a3 a4 d g u
#> par 3.06 0 0.501 0 -5.002 0.1 1
#>
#> $Item.7
#> a1 a2 a3 a4 d g u
#> par 1.121 0.839 0 0 1.373 0.1 1
#>
#> $Item.8
#> a1 a2 a3 a4 d g u
#> par 1.956 0 1.443 0 -3.772 0.1 1
#>
#> $Item.9
#> a1 a2 a3 a4 d g u
#> par 0.512 1.236 0 0 2.484 0.1 1
#>
#> $Item.10
#> a1 a2 a3 a4 d g u
#> par 1.68 1.506 0 0 -1.031 0.1 1
#>
#> $Item.11
#> a1 a2 a3 a4 d g u
#> par 1.655 0.842 0 0 5.441 0.1 1
#>
#> $Item.12
#> a1 a2 a3 a4 d g u
#> par 0.129 0 0 0.364 -0.641 0.1 1
#>
#> $Item.13
#> a1 a2 a3 a4 d g u
#> par 1.183 0.477 0 0 0.679 0.1 1
#>
#> $Item.14
#> a1 a2 a3 a4 d g u
#> par 1.125 0 0 1.058 1.164 0.1 1
#>
#> $Item.15
#> a1 a2 a3 a4 d g u
#> par 1.435 0.317 0 0 1.863 0.1 1
#>
#> $Item.16
#> a1 a2 a3 a4 d g u
#> par 0.95 0 0.573 0 -0.783 0.1 1
#>
#> $Item.17
#> a1 a2 a3 a4 d g u
#> par 1.547 0.059 0 0 4.112 0.1 1
#>
#> $Item.18
#> a1 a2 a3 a4 d g u
#> par 2.731 0.094 0 0 -1.808 0.1 1
#>
#> $Item.19
#> a1 a2 a3 a4 d g u
#> par 0.918 0 0 0.101 -0.001 0.1 1
#>
#> $Item.20
#> a1 a2 a3 a4 d g u
#> par 1.456 0 0 0.593 2.501 0.1 1
#>
#> $Item.21
#> a1 a2 a3 a4 d g u
#> par 0.596 0.493 0 0 2.49 0.1 1
#>
#> $Item.22
#> a1 a2 a3 a4 d g u
#> par 1.554 -0.242 0 0 3.428 0.1 1
#>
#> $Item.23
#> a1 a2 a3 a4 d g u
#> par 0.908 0 0 0.766 -1.488 0.1 1
#>
#> $Item.24
#> a1 a2 a3 a4 d g u
#> par 1.379 0.001 0 0 1.132 0.1 1
#>
#> $Item.25
#> a1 a2 a3 a4 d g u
#> par 1.03 0 0 1.094 -1.164 0.1 1
#>
#> $Item.26
#> a1 a2 a3 a4 d g u
#> par 1.985 0 0 0.747 -0.663 0.1 1
#>
#> $Item.27
#> a1 a2 a3 a4 d g u
#> par 1.909 0.348 0 0 2.642 0.1 1
#>
#> $Item.28
#> a1 a2 a3 a4 d g u
#> par 1.213 0 0 0.142 -0.097 0.1 1
#>
#> $Item.29
#> a1 a2 a3 a4 d g u
#> par 1.938 0 2.339 0 -2.209 0.1 1
#>
#> $Item.30
#> a1 a2 a3 a4 d g u
#> par 0.479 0 0 -0.128 -0.527 0.1 1
#>
#> $Item.31
#> a1 a2 a3 a4 d g u
#> par 3.173 -0.82 0 0 3.316 0.1 1
#>
#> $Item.32
#> a1 a2 a3 a4 d g u
#> par 0.534 0 -0.053 0 -2.786 0.1 1
#>
#> $GroupPars
#> MEAN_1 MEAN_2 MEAN_3 MEAN_4 COV_11 COV_21 COV_31 COV_41 COV_22 COV_32
#> par 0 0 0 0 1 0 0 0 1 0
#> COV_42 COV_33 COV_43 COV_44
#> par 0 1 0 1
#>
anova(mod1, mod2)
#> AIC SABIC HQ BIC logLik X2 df p
#> mod1 19062.10 19179.44 19226.42 19484.21 -9435.052
#> mod2 19009.55 19126.88 19173.87 19431.65 -9408.775 52.553 0 NaN
## don't estimate specific factor for item 32
specific[32] <- NA
mod3 <- bfactor(data, specific)
anova(mod3, mod1)
#> AIC SABIC HQ BIC logLik X2 df p
#> mod3 19060.12 19176.23 19222.73 19477.83 -9435.062
#> mod1 19062.10 19179.44 19226.42 19484.21 -9435.052 0.02 1 0.886
# same, but with syntax (not run)
specific3 <- "S1 = 7,9,10,11,13,15,17,18,21,22,24,27,31
S2 = 1,3,6,8,16,29
S3 = 2,4,5,12,14,19,20,23,25,26,28,30"
# mod3b <- bfactor(data, specific3)
# anova(mod3b)
#########
# mixed itemtype example
# simulate data
a <- matrix(c(
1,0.5,NA,
1,0.5,NA,
1,0.5,NA,
1,0.5,NA,
1,0.5,NA,
1,0.5,NA,
1,0.5,NA,
1,NA,0.5,
1,NA,0.5,
1,NA,0.5,
1,NA,0.5,
1,NA,0.5,
1,NA,0.5,
1,NA,0.5),ncol=3,byrow=TRUE)
d <- matrix(c(
-1.0,NA,NA,
-1.5,NA,NA,
1.5,NA,NA,
0.0,NA,NA,
2.5,1.0,-1,
3.0,2.0,-0.5,
3.0,2.0,-0.5,
3.0,2.0,-0.5,
2.5,1.0,-1,
2.0,0.0,NA,
-1.0,NA,NA,
-1.5,NA,NA,
1.5,NA,NA,
0.0,NA,NA),ncol=3,byrow=TRUE)
items <- rep('2PL', 14)
items[5:10] <- 'graded'
sigma <- diag(3)
dataset <- simdata(a,d,5000,itemtype=items,sigma=sigma)
itemstats(dataset)
#> $overall
#> N mean_total.score sd_total.score ave.r sd.r alpha SEM.alpha
#> 5000 15.116 4.495 0.175 0.034 0.733 2.323
#>
#> $itemstats
#> N mean sd total.r total.r_if_rm alpha_if_rm
#> Item_1 5000 0.311 0.463 0.434 0.345 0.720
#> Item_2 5000 0.229 0.420 0.386 0.302 0.724
#> Item_3 5000 0.770 0.421 0.378 0.293 0.724
#> Item_4 5000 0.493 0.500 0.455 0.360 0.718
#> Item_5 5000 1.893 0.975 0.585 0.414 0.711
#> Item_6 5000 2.150 0.883 0.537 0.374 0.716
#> Item_7 5000 2.168 0.881 0.538 0.376 0.715
#> Item_8 5000 2.138 0.889 0.549 0.388 0.714
#> Item_9 5000 1.857 0.978 0.586 0.413 0.711
#> Item_10 5000 1.327 0.746 0.523 0.387 0.713
#> Item_11 5000 0.304 0.460 0.427 0.338 0.721
#> Item_12 5000 0.234 0.424 0.403 0.320 0.723
#> Item_13 5000 0.752 0.432 0.417 0.333 0.722
#> Item_14 5000 0.491 0.500 0.457 0.362 0.718
#>
#> $proportions
#> 0 1 2 3
#> Item_1 0.689 0.311 NA NA
#> Item_2 0.771 0.229 NA NA
#> Item_3 0.230 0.770 NA NA
#> Item_4 0.507 0.493 NA NA
#> Item_5 0.114 0.194 0.378 0.314
#> Item_6 0.079 0.089 0.434 0.397
#> Item_7 0.079 0.083 0.431 0.408
#> Item_8 0.081 0.093 0.433 0.393
#> Item_9 0.115 0.214 0.370 0.301
#> Item_10 0.168 0.337 0.495 NA
#> Item_11 0.696 0.304 NA NA
#> Item_12 0.766 0.234 NA NA
#> Item_13 0.248 0.752 NA NA
#> Item_14 0.509 0.491 NA NA
#>
specific <- "S1 = 1-7
S2 = 8-14"
simmod <- bfactor(dataset, specific)
coef(simmod, simplify=TRUE)
#> $items
#> a1 a2 a3 d g u d1 d2 d3
#> Item_1 1.032 0.505 0.000 -1.003 0 1 NA NA NA
#> Item_2 0.953 0.474 0.000 -1.477 0 1 NA NA NA
#> Item_3 0.868 0.494 0.000 1.442 0 1 NA NA NA
#> Item_4 1.002 0.406 0.000 -0.037 0 1 NA NA NA
#> Item_5 1.040 0.578 0.000 NA NA NA 2.536 1.029 -0.998
#> Item_6 0.963 0.465 0.000 NA NA NA 2.899 1.928 -0.514
#> Item_7 0.969 0.504 0.000 NA NA NA 2.929 2.009 -0.462
#> Item_8 1.011 0.000 0.680 NA NA NA 2.993 1.974 -0.563
#> Item_9 1.023 0.000 0.469 NA NA NA 2.471 0.888 -1.054
#> Item_10 1.002 0.000 0.461 NA NA NA 1.951 -0.023 NA
#> Item_11 1.001 0.000 0.543 -1.041 0 1 NA NA NA
#> Item_12 1.039 0.000 0.603 -1.502 0 1 NA NA NA
#> Item_13 1.033 0.000 0.429 1.377 0 1 NA NA NA
#> Item_14 1.017 0.000 0.401 -0.046 0 1 NA NA NA
#>
#> $means
#> G S1 S2
#> 0 0 0
#>
#> $cov
#> G S1 S2
#> G 1 0 0
#> S1 0 1 0
#> S2 0 0 1
#>
#########
# General testlet response model (Wainer, 2007)
# simulate data
set.seed(1234)
a <- matrix(0, 12, 4)
a[,1] <- rlnorm(12, .2, .3)
ind <- 1
for(i in 1:3){
a[ind:(ind+3),i+1] <- a[ind:(ind+3),1]
ind <- ind+4
}
print(a)
#> [,1] [,2] [,3] [,4]
#> [1,] 0.8503394 0.8503394 0.000000 0.0000000
#> [2,] 1.3274088 1.3274088 0.000000 0.0000000
#> [3,] 1.6910208 1.6910208 0.000000 0.0000000
#> [4,] 0.6042850 0.6042850 0.000000 0.0000000
#> [5,] 1.3892130 0.0000000 1.389213 0.0000000
#> [6,] 1.4216480 0.0000000 1.421648 0.0000000
#> [7,] 1.0279618 0.0000000 1.027962 0.0000000
#> [8,] 1.0366667 0.0000000 1.036667 0.0000000
#> [9,] 1.0311394 0.0000000 0.000000 1.0311394
#> [10,] 0.9351846 0.0000000 0.000000 0.9351846
#> [11,] 1.0584888 0.0000000 0.000000 1.0584888
#> [12,] 0.9052755 0.0000000 0.000000 0.9052755
d <- rnorm(12, 0, .5)
sigma <- diag(c(1, .5, 1, .5))
dataset <- simdata(a,d,2000,itemtype=rep('2PL', 12),sigma=sigma)
itemstats(dataset)
#> $overall
#> N mean_total.score sd_total.score ave.r sd.r alpha SEM.alpha
#> 2000 6 2.929 0.175 0.068 0.717 1.558
#>
#> $itemstats
#> N mean sd total.r total.r_if_rm alpha_if_rm
#> Item_1 2000 0.426 0.495 0.438 0.287 0.708
#> Item_2 2000 0.502 0.500 0.560 0.425 0.689
#> Item_3 2000 0.571 0.495 0.575 0.445 0.686
#> Item_4 2000 0.502 0.500 0.383 0.224 0.716
#> Item_5 2000 0.464 0.499 0.549 0.413 0.690
#> Item_6 2000 0.436 0.496 0.561 0.428 0.688
#> Item_7 2000 0.440 0.497 0.500 0.356 0.698
#> Item_8 2000 0.693 0.462 0.474 0.339 0.701
#> Item_9 2000 0.511 0.500 0.481 0.334 0.701
#> Item_10 2000 0.456 0.498 0.465 0.316 0.704
#> Item_11 2000 0.458 0.498 0.459 0.309 0.705
#> Item_12 2000 0.540 0.498 0.475 0.327 0.702
#>
#> $proportions
#> 0 1
#> Item_1 0.575 0.426
#> Item_2 0.498 0.502
#> Item_3 0.430 0.571
#> Item_4 0.498 0.502
#> Item_5 0.536 0.464
#> Item_6 0.564 0.436
#> Item_7 0.559 0.440
#> Item_8 0.308 0.693
#> Item_9 0.488 0.511
#> Item_10 0.543 0.456
#> Item_11 0.541 0.458
#> Item_12 0.460 0.540
#>
# estimate by applying constraints and freeing the latent variances
specific <- "S1 = 1-4
S2 = 5-8
S3 = 9-12"
model <- "G = 1-12
CONSTRAIN = (1, a1, a2), (2, a1, a2), (3, a1, a2), (4, a1, a2),
(5, a1, a3), (6, a1, a3), (7, a1, a3), (8, a1, a3),
(9, a1, a4), (10, a1, a4), (11, a1, a4), (12, a1, a4)
COV = S1*S1, S2*S2, S3*S3"
simmod <- bfactor(dataset, specific, model)
coef(simmod, simplify=TRUE)
#> $items
#> a1 a2 a3 a4 d g u
#> Item_1 0.794 0.794 0.000 0.000 -0.359 0 1
#> Item_2 1.544 1.544 0.000 0.000 0.011 0 1
#> Item_3 1.762 1.762 0.000 0.000 0.479 0 1
#> Item_4 0.544 0.544 0.000 0.000 0.011 0 1
#> Item_5 1.386 0.000 1.386 0.000 -0.244 0 1
#> Item_6 1.497 0.000 1.497 0.000 -0.449 0 1
#> Item_7 0.853 0.000 0.853 0.000 -0.312 0 1
#> Item_8 0.953 0.000 0.953 0.000 1.101 0 1
#> Item_9 0.981 0.000 0.000 0.981 0.058 0 1
#> Item_10 0.913 0.000 0.000 0.913 -0.217 0 1
#> Item_11 0.868 0.000 0.000 0.868 -0.204 0 1
#> Item_12 0.966 0.000 0.000 0.966 0.206 0 1
#>
#> $means
#> G S1 S2 S3
#> 0 0 0 0
#>
#> $cov
#> G S1 S2 S3
#> G 1 0.000 0.000 0.000
#> S1 0 0.452 0.000 0.000
#> S2 0 0.000 1.135 0.000
#> S3 0 0.000 0.000 0.432
#>
# Constrained testlet model (Bradlow, 1999)
model2 <- "G = 1-12
CONSTRAIN = (1, a1, a2), (2, a1, a2), (3, a1, a2), (4, a1, a2),
(5, a1, a3), (6, a1, a3), (7, a1, a3), (8, a1, a3),
(9, a1, a4), (10, a1, a4), (11, a1, a4), (12, a1, a4),
(GROUP, COV_22, COV_33, COV_44)
COV = S1*S1, S2*S2, S3*S3"
simmod2 <- bfactor(dataset, specific, model2)
coef(simmod2, simplify=TRUE)
#> $items
#> a1 a2 a3 a4 d g u
#> Item_1 0.744 0.744 0.000 0.000 -0.360 0 1
#> Item_2 1.453 1.453 0.000 0.000 0.010 0 1
#> Item_3 1.664 1.664 0.000 0.000 0.482 0 1
#> Item_4 0.509 0.509 0.000 0.000 0.011 0 1
#> Item_5 1.541 0.000 1.541 0.000 -0.241 0 1
#> Item_6 1.670 0.000 1.670 0.000 -0.445 0 1
#> Item_7 0.968 0.000 0.968 0.000 -0.313 0 1
#> Item_8 1.075 0.000 1.075 0.000 1.098 0 1
#> Item_9 0.927 0.000 0.000 0.927 0.059 0 1
#> Item_10 0.854 0.000 0.000 0.854 -0.218 0 1
#> Item_11 0.813 0.000 0.000 0.813 -0.205 0 1
#> Item_12 0.908 0.000 0.000 0.908 0.207 0 1
#>
#> $means
#> G S1 S2 S3
#> 0 0 0 0
#>
#> $cov
#> G S1 S2 S3
#> G 1 0.000 0.000 0.000
#> S1 0 0.667 0.000 0.000
#> S2 0 0.000 0.667 0.000
#> S3 0 0.000 0.000 0.667
#>
anova(simmod2, simmod)
#> AIC SABIC HQ BIC logLik X2 df p
#> simmod2 30256.59 30317.19 30308.00 30396.61 -15103.3
#> simmod 30248.79 30314.24 30304.32 30400.02 -15097.4 11.795 2 0.003
#########
# Two-tier model
# simulate data
set.seed(1234)
a <- matrix(c(
0,1,0.5,NA,NA,
0,1,0.5,NA,NA,
0,1,0.5,NA,NA,
0,1,0.5,NA,NA,
0,1,0.5,NA,NA,
0,1,NA,0.5,NA,
0,1,NA,0.5,NA,
0,1,NA,0.5,NA,
1,0,NA,0.5,NA,
1,0,NA,0.5,NA,
1,0,NA,0.5,NA,
1,0,NA,NA,0.5,
1,0,NA,NA,0.5,
1,0,NA,NA,0.5,
1,0,NA,NA,0.5,
1,0,NA,NA,0.5),ncol=5,byrow=TRUE)
d <- matrix(rnorm(16))
items <- rep('2PL', 16)
sigma <- diag(5)
sigma[1,2] <- sigma[2,1] <- .4
dataset <- simdata(a,d,2000,itemtype=items,sigma=sigma)
itemstats(dataset)
#> $overall
#> N mean_total.score sd_total.score ave.r sd.r alpha SEM.alpha
#> 2000 7.086 3.077 0.108 0.058 0.662 1.79
#>
#> $itemstats
#> N mean sd total.r total.r_if_rm alpha_if_rm
#> Item_1 2000 0.288 0.453 0.378 0.241 0.650
#> Item_2 2000 0.571 0.495 0.422 0.276 0.646
#> Item_3 2000 0.705 0.456 0.381 0.245 0.650
#> Item_4 2000 0.133 0.340 0.289 0.183 0.656
#> Item_5 2000 0.601 0.490 0.393 0.246 0.650
#> Item_6 2000 0.587 0.492 0.419 0.274 0.646
#> Item_7 2000 0.379 0.485 0.444 0.304 0.642
#> Item_8 2000 0.378 0.485 0.400 0.256 0.649
#> Item_9 2000 0.386 0.487 0.392 0.246 0.650
#> Item_10 2000 0.322 0.467 0.400 0.261 0.648
#> Item_11 2000 0.402 0.490 0.455 0.315 0.640
#> Item_12 2000 0.318 0.466 0.414 0.278 0.646
#> Item_13 2000 0.368 0.482 0.423 0.281 0.645
#> Item_14 2000 0.498 0.500 0.424 0.277 0.646
#> Item_15 2000 0.669 0.471 0.394 0.254 0.649
#> Item_16 2000 0.482 0.500 0.444 0.300 0.642
#>
#> $proportions
#> 0 1
#> Item_1 0.713 0.288
#> Item_2 0.430 0.571
#> Item_3 0.295 0.705
#> Item_4 0.867 0.133
#> Item_5 0.400 0.601
#> Item_6 0.413 0.587
#> Item_7 0.621 0.379
#> Item_8 0.622 0.378
#> Item_9 0.614 0.386
#> Item_10 0.678 0.322
#> Item_11 0.598 0.402
#> Item_12 0.681 0.318
#> Item_13 0.632 0.368
#> Item_14 0.502 0.498
#> Item_15 0.330 0.669
#> Item_16 0.518 0.482
#>
specific <- "S1 = 1-5
S2 = 6-11
S3 = 12-16"
model <- '
G1 = 1-8
G2 = 9-16
COV = G1*G2'
# quadpts dropped for faster estimation, but not as precise
simmod <- bfactor(dataset, specific, model, quadpts = 9, TOL = 1e-3)
coef(simmod, simplify=TRUE)
#> $items
#> a1 a2 a3 a4 a5 d g u
#> Item_1 0.965 0.000 0.385 0.000 0.000 -1.100 0 1
#> Item_2 1.076 0.000 0.550 0.000 0.000 0.363 0 1
#> Item_3 0.898 0.000 0.592 0.000 0.000 1.068 0 1
#> Item_4 0.896 0.000 0.710 0.000 0.000 -2.293 0 1
#> Item_5 0.892 0.000 0.848 0.000 0.000 0.526 0 1
#> Item_6 1.013 0.000 0.000 0.413 0.000 0.435 0 1
#> Item_7 1.162 0.000 0.000 0.451 0.000 -0.639 0 1
#> Item_8 0.945 0.000 0.000 0.609 0.000 -0.623 0 1
#> Item_9 0.000 0.831 0.000 0.371 0.000 -0.544 0 1
#> Item_10 0.000 0.925 0.000 0.610 0.000 -0.926 0 1
#> Item_11 0.000 1.142 0.000 0.495 0.000 -0.517 0 1
#> Item_12 0.000 0.978 0.000 0.000 0.634 -0.964 0 1
#> Item_13 0.000 1.108 0.000 0.000 0.437 -0.694 0 1
#> Item_14 0.000 1.004 0.000 0.000 0.321 -0.012 0 1
#> Item_15 0.000 0.916 0.000 0.000 0.758 0.897 0 1
#> Item_16 0.000 1.020 0.000 0.000 0.650 -0.096 0 1
#>
#> $means
#> G1 G2 S1 S2 S3
#> 0 0 0 0 0
#>
#> $cov
#> G1 G2 S1 S2 S3
#> G1 1.000 0.412 0 0 0
#> G2 0.412 1.000 0 0 0
#> S1 0.000 0.000 1 0 0
#> S2 0.000 0.000 0 1 0
#> S3 0.000 0.000 0 0 1
#>
summary(simmod)
#> G1 G2 S1 S2 S3 h2
#> Item_1 0.484 0.000 0.193 0.000 0.000 0.271
#> Item_2 0.516 0.000 0.263 0.000 0.000 0.335
#> Item_3 0.446 0.000 0.294 0.000 0.000 0.285
#> Item_4 0.437 0.000 0.346 0.000 0.000 0.311
#> Item_5 0.425 0.000 0.404 0.000 0.000 0.343
#> Item_6 0.501 0.000 0.000 0.204 0.000 0.293
#> Item_7 0.551 0.000 0.000 0.214 0.000 0.349
#> Item_8 0.463 0.000 0.000 0.299 0.000 0.304
#> Item_9 0.000 0.431 0.000 0.192 0.000 0.222
#> Item_10 0.000 0.456 0.000 0.300 0.000 0.298
#> Item_11 0.000 0.541 0.000 0.235 0.000 0.348
#> Item_12 0.000 0.474 0.000 0.000 0.307 0.319
#> Item_13 0.000 0.533 0.000 0.000 0.210 0.329
#> Item_14 0.000 0.501 0.000 0.000 0.160 0.277
#> Item_15 0.000 0.441 0.000 0.000 0.365 0.328
#> Item_16 0.000 0.488 0.000 0.000 0.311 0.336
#>
#> SS loadings: 1.839 1.88 0.476 0.359 0.395
#> Proportion Var: 0.115 0.118 0.03 0.022 0.025
#>
#> Factor correlations:
#>
#> G1 G2 S1 S2 S3
#> G1 1.000
#> G2 0.412 1
#> S1 0.000 0 1
#> S2 0.000 0 0 1
#> S3 0.000 0 0 0 1
itemfit(simmod, QMC=TRUE)
#> item S_X2 df.S_X2 RMSEA.S_X2 p.S_X2
#> 1 Item_1 7.103 9 0.000 0.626
#> 2 Item_2 13.326 10 0.013 0.206
#> 3 Item_3 8.332 9 0.000 0.501
#> 4 Item_4 8.531 10 0.000 0.577
#> 5 Item_5 7.170 10 0.000 0.709
#> 6 Item_6 3.967 10 0.000 0.949
#> 7 Item_7 8.350 10 0.000 0.595
#> 8 Item_8 16.010 10 0.017 0.099
#> 9 Item_9 17.529 10 0.019 0.063
#> 10 Item_10 12.058 10 0.010 0.281
#> 11 Item_11 13.567 10 0.013 0.194
#> 12 Item_12 13.907 9 0.017 0.126
#> 13 Item_13 11.144 10 0.008 0.346
#> 14 Item_14 7.852 10 0.000 0.643
#> 15 Item_15 14.142 9 0.017 0.117
#> 16 Item_16 5.926 10 0.000 0.821
M2(simmod, QMC=TRUE)
#> M2 df p RMSEA RMSEA_5 RMSEA_95 SRMSR TLI CFI
#> stats 86.28163 87 0.5015988 0 0 0.01201603 0.01662365 1.000285 1
residuals(simmod, QMC=TRUE)
#> LD matrix (lower triangle) and standardized residual correlations (upper triangle)
#>
#> Upper triangle summary:
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> -0.046 -0.011 -0.002 -0.001 0.012 0.041
#>
#> Item_1 Item_2 Item_3 Item_4 Item_5 Item_6 Item_7 Item_8 Item_9 Item_10
#> Item_1 -0.011 -0.015 -0.007 0.016 0.003 0.006 -0.002 0.004 -0.009
#> Item_2 0.263 -0.002 0.005 -0.001 0.016 -0.022 0.021 -0.003 -0.006
#> Item_3 0.441 0.008 0.014 -0.001 0.029 -0.017 -0.011 -0.021 -0.028
#> Item_4 0.086 0.054 0.376 -0.014 -0.023 0.021 -0.004 0.020 -0.040
#> Item_5 0.514 0.004 0.003 0.386 -0.028 0.011 0.014 -0.021 0.013
#> Item_6 0.015 0.483 1.630 1.038 1.588 -0.022 -0.009 0.031 -0.004
#> Item_7 0.077 0.996 0.590 0.852 0.258 0.992 -0.007 -0.004 0.013
#> Item_8 0.012 0.858 0.264 0.039 0.377 0.154 0.094 -0.020 0.008
#> Item_9 0.033 0.017 0.863 0.803 0.890 1.974 0.037 0.808 0.001
#> Item_10 0.157 0.084 1.528 3.158 0.360 0.038 0.330 0.128 0.001
#> Item_11 0.510 0.125 2.195 0.231 0.004 0.215 1.049 0.004 0.031 0.754
#> Item_12 2.017 0.253 1.865 0.388 0.005 0.417 0.074 0.090 0.443 0.042
#> Item_13 0.470 2.122 0.125 0.271 0.881 0.264 0.310 4.304 0.009 0.059
#> Item_14 0.101 1.546 0.165 0.006 0.296 0.004 1.672 0.765 3.341 0.066
#> Item_15 0.822 0.257 0.011 0.442 0.443 0.113 0.526 0.297 2.306 0.044
#> Item_16 0.097 0.627 1.486 0.127 0.445 0.011 0.732 0.061 0.007 0.944
#> Item_11 Item_12 Item_13 Item_14 Item_15 Item_16
#> Item_1 -0.016 -0.032 0.015 -0.007 0.020 -0.007
#> Item_2 0.008 0.011 -0.033 -0.028 -0.011 0.018
#> Item_3 0.033 0.031 0.008 -0.009 -0.002 0.027
#> Item_4 -0.011 0.014 -0.012 -0.002 -0.015 -0.008
#> Item_5 -0.001 0.002 -0.021 -0.012 0.015 0.015
#> Item_6 0.010 0.014 -0.011 0.001 -0.008 0.002
#> Item_7 0.023 0.006 0.012 0.029 -0.016 0.019
#> Item_8 -0.001 0.007 -0.046 -0.020 0.012 0.006
#> Item_9 0.004 -0.015 -0.002 0.041 -0.034 -0.002
#> Item_10 -0.019 0.005 0.005 0.006 -0.005 0.022
#> Item_11 0.009 0.000 -0.025 0.039 -0.011
#> Item_12 0.149 -0.008 0.013 -0.002 -0.009
#> Item_13 0.000 0.140 0.013 -0.012 0.003
#> Item_14 1.269 0.338 0.336 -0.007 -0.023
#> Item_15 3.061 0.010 0.284 0.086 0.010
#> Item_16 0.250 0.153 0.015 1.026 0.190
# }